The Lambda Variant in Argentina: Analyzing the Evolution and Spread of SARS-CoV-2 Lineage C.37
- PMID: 37376681
- PMCID: PMC10305049
- DOI: 10.3390/v15061382
The Lambda Variant in Argentina: Analyzing the Evolution and Spread of SARS-CoV-2 Lineage C.37
Abstract
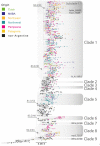
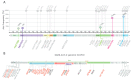
The second wave of COVID-19 occurred in South America in early 2021 and was mainly driven by Gamma and Lambda variants. In this study, we aimed to describe the emergence and local genomic diversity of the SARS-CoV-2 Lambda variant in Argentina, from its initial entry into the country until its detection ceased. Molecular surveillance was conducted on 9356 samples from Argentina between October 2020 and April 2022, and sequencing, phylogenetic, and phylogeographic analyses were performed. Our findings revealed that the Lambda variant was first detected in Argentina in January 2021 and steadily increased in frequency until it peaked in April 2021, with continued detection throughout the year. Phylodynamic analyses showed that at least 18 introductions of the Lambda variant into the country occurred, with nine of them having evidence of onward local transmission. The spatial--temporal reconstruction showed that Argentine clades were associated with Lambda sequences from Latin America and suggested an initial diversification in the Metropolitan Area of Buenos Aires before spreading to other regions in Argentina. Genetic analyses of genome sequences allowed us to describe the mutational patterns of the Argentine Lambda sequences and detect the emergence of rare mutations in an immunocompromised patient. Our study highlights the importance of genomic surveillance in identifying the introduction and geographical distribution of the SARS-CoV-2 Lambda variant, as well as in monitoring the emergence of mutations that could be involved in the evolutionary leaps that characterize variants of concern.
Keywords: Lambda; SARS-CoV-2; South America; evolution; phylodynamic; variants.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




References
-
- World Health Organization Tracking SARS-CoV-2 Variants. [(accessed on 25 February 2023)];2021 Available online: https://Www.Who.Int/En/Activities/Tracking-SARS-CoV-2-Variants/
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

