Identification of immunosuppressive signature subtypes and prognostic risk signatures in triple-negative breast cancer
- PMID: 37377907
- PMCID: PMC10292819
- DOI: 10.3389/fonc.2023.1108472
Identification of immunosuppressive signature subtypes and prognostic risk signatures in triple-negative breast cancer
Abstract
Purpose: Immune checkpoint blockade (ICB) therapy has transformed the treatment of triple-negative breast cancer (TNBC) in recent years. However, some TNBC patients with high programmed death-ligand 1 (PD-L1) expression levels develop immune checkpoint resistance. Hence, there is an urgent need to characterize the immunosuppressive tumor microenvironment and identify biomarkers to construct prognostic models of patient survival outcomes in order to understand biological mechanisms operating within the tumor microenvironment.
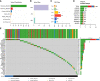
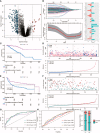
Patients and methods: RNA sequence (RNA-seq) data from 303 TNBC samples were analyzed using an unsupervised cluster analysis approach to reveal distinctive cellular gene expression patterns within the TNBC tumor microenvironment (TME). A panel of T cell exhaustion signatures, immunosuppressive cell subtypes and clinical features were correlated with the immunotherapeutic response, as assessed according to gene expression patterns. The test dataset was then used to confirm the occurrence of immune depletion status and prognostic features and to formulate clinical treatment recommendations. Concurrently, a reliable risk prediction model and clinical treatment strategy were proposed based on TME immunosuppressive signature differences between TNBC patients with good versus poor survival status and other clinical prognostic factors.
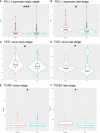
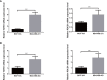
Results: Significantly enriched TNBC microenvironment T cell depletion signatures were detected in the analyzed RNA-seq data. A high proportion of certain immunosuppressive cell subtypes, 9 inhibitory checkpoints and enhanced anti-inflammatory cytokine expression profiles were noted in 21.4% of TNBC patients that led to the designation of this group of immunosuppressed patients as the immune depletion class (IDC). Although IDC group TNBC samples contained tumor-infiltrating lymphocytes present at high densities, IDC patient prognosis was poor. Notably, PD-L1 expression was relatively elevated in IDC patients that indicated their cancers were resistant to ICB treatment. Based on these findings, a set of gene expression signatures predicting IDC group PD-L1 resistance was identified then used to develop risk models for use in predicting clinical therapeutic outcomes.
Conclusion: A novel TNBC immunosuppressive tumor microenvironment subtype associated with strong PD-L1 expression and possible resistance to ICB treatment was identified. This comprehensive gene expression pattern may provide fresh insights into drug resistance mechanisms for use in optimizing immunotherapeutic approaches for TNBC patients.
Keywords: TNBC; immunogenomics; immunosuppressive cytokines; t cell exhaustion; tumor microenvironment.
Copyright © 2023 Ding, Wang, Fan, Tian, Wang, Qin, Su and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources
Research Materials

