Plasma protein biomarkers for early prediction of lung cancer
- PMID: 37379654
- PMCID: PMC10320232
- DOI: 10.1016/j.ebiom.2023.104686
Plasma protein biomarkers for early prediction of lung cancer
Abstract
Background: Individual plasma proteins have been identified as minimally invasive biomarkers for lung cancer diagnosis with potential utility in early detection. Plasma proteomes provide insight into contributing biological factors; we investigated their potential for future lung cancer prediction.
Methods: The Olink® Explore-3072 platform quantitated 2941 proteins in 496 Liverpool Lung Project plasma samples, including 131 cases taken 1-10 years prior to diagnosis, 237 controls, and 90 subjects at multiple times. 1112 proteins significantly associated with haemolysis were excluded. Feature selection with bootstrapping identified differentially expressed proteins, subsequently modelled for lung cancer prediction and validated in UK Biobank data.
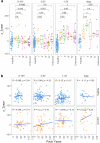
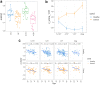
Findings: For samples 1-3 years pre-diagnosis, 240 proteins were significantly different in cases; for 1-5 year samples, 117 of these and 150 further proteins were identified, mapping to significantly different pathways. Four machine learning algorithms gave median AUCs of 0.76-0.90 and 0.73-0.83 for the 1-3 year and 1-5 year proteins respectively. External validation gave AUCs of 0.75 (1-3 year) and 0.69 (1-5 year), with AUC 0.7 up to 12 years prior to diagnosis. The models were independent of age, smoking duration, cancer histology and the presence of COPD.
Interpretation: The plasma proteome provides biomarkers which may be used to identify those at greatest risk of lung cancer. The proteins and the pathways are different when lung cancer is more imminent, indicating that both biomarkers of inherent risk and biomarkers associated with presence of early lung cancer may be identified.
Funding: Janssen Pharmaceuticals Research Collaboration Award; Roy Castle Lung Cancer Foundation.
Keywords: Early-detection; Lung cancer prediction; Plasma; Proteins; Proteomics.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests This work was funded through a Research Collaboration Agreement between Janssen Pharmaceuticals and the University of Liverpool: MD & JKF received research funding from Janssen Pharmaceuticals (a Johnson & Johnson company). TS, HA, LH & RY are employees of Johnson & Johnson, the company has filed a patent to on use of plasma protein biomarkers in lung cancer interception. TL declares no conflict of interest. Both parties shared responsibility for: study design; collection, analysis and interpretation of experimental data; writing the report and the decision to publish.
Figures


References
-
- Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2021;71(3):209–249. - PubMed
-
- Miller K.D., Nogueira L., Mariotto A.B., et al. Cancer treatment and survivorship statistics, 2019. CA A Cancer J Clin. 2019;69(5):363–385. - PubMed
-
- Nicholson A.G., Chansky K., Crowley J., et al. The international association for the study of lung cancer lung cancer staging project: proposals for the revision of the clinical and pathologic staging of small cell lung cancer in the forthcoming eighth edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11(3):300–311. - PubMed
-
- de Koning H.J., van der Aalst C.M., de Jong P.A., et al. Reduced lung-cancer mortality with volume CT screening in a randomized trial. N Engl J Med. 2020;382(6):503–513. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

