Phase separation of FSP1 promotes ferroptosis
- PMID: 37380771
- PMCID: PMC10338336
- DOI: 10.1038/s41586-023-06255-6
Phase separation of FSP1 promotes ferroptosis
Abstract
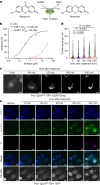
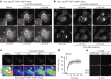
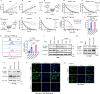
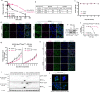
Ferroptosis is evolving as a highly promising approach to combat difficult-to-treat tumour entities including therapy-refractory and dedifferentiating cancers1-3. Recently, ferroptosis suppressor protein-1 (FSP1), along with extramitochondrial ubiquinone or exogenous vitamin K and NAD(P)H/H+ as an electron donor, has been identified as the second ferroptosis-suppressing system, which efficiently prevents lipid peroxidation independently of the cyst(e)ine-glutathione (GSH)-glutathione peroxidase 4 (GPX4) axis4-6. To develop FSP1 inhibitors as next-generation therapeutic ferroptosis inducers, here we performed a small molecule library screen and identified the compound class of 3-phenylquinazolinones (represented by icFSP1) as potent FSP1 inhibitors. We show that icFSP1, unlike iFSP1, the first described on-target FSP1 inhibitor5, does not competitively inhibit FSP1 enzyme activity, but instead triggers subcellular relocalization of FSP1 from the membrane and FSP1 condensation before ferroptosis induction, in synergism with GPX4 inhibition. icFSP1-induced FSP1 condensates show droplet-like properties consistent with phase separation, an emerging and widespread mechanism to modulate biological activity7. N-terminal myristoylation, distinct amino acid residues and intrinsically disordered, low-complexity regions in FSP1 were identified to be essential for FSP1-dependent phase separation in cells and in vitro. We further demonstrate that icFSP1 impairs tumour growth and induces FSP1 condensates in tumours in vivo. Hence, our results suggest that icFSP1 exhibits a unique mechanism of action and synergizes with ferroptosis-inducing agents to potentiate the ferroptotic cell death response, thus providing a rationale for targeting FSP1-dependent phase separation as an efficient anti-cancer therapy.
© 2023. The Author(s).
Conflict of interest statement
M.C., B.P. and T.N. hold patents for some of the compounds described herein, and M.C. and B.P. are co-founders and shareholders of ROSCUE Therapeutics GmbH. The other authors declare no competing interests.
Figures









Comment in
-
FSP1 in cancer: not just a phase.Nat Rev Cancer. 2023 Sep;23(9):578. doi: 10.1038/s41568-023-00607-0. Nat Rev Cancer. 2023. PMID: 37491632 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

