Continuous synthesis of E. coli genome sections and Mb-scale human DNA assembly
- PMID: 37380776
- PMCID: PMC7614783
- DOI: 10.1038/s41586-023-06268-1
Continuous synthesis of E. coli genome sections and Mb-scale human DNA assembly
Abstract
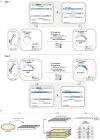
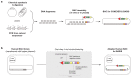
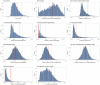
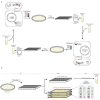
Whole-genome synthesis provides a powerful approach for understanding and expanding organism function1-3. To build large genomes rapidly, scalably and in parallel, we need (1) methods for assembling megabases of DNA from shorter precursors and (2) strategies for rapidly and scalably replacing the genomic DNA of organisms with synthetic DNA. Here we develop bacterial artificial chromosome (BAC) stepwise insertion synthesis (BASIS)-a method for megabase-scale assembly of DNA in Escherichia coli episomes. We used BASIS to assemble 1.1 Mb of human DNA containing numerous exons, introns, repetitive sequences, G-quadruplexes, and long and short interspersed nuclear elements (LINEs and SINEs). BASIS provides a powerful platform for building synthetic genomes for diverse organisms. We also developed continuous genome synthesis (CGS)-a method for continuously replacing sequential 100 kb stretches of the E. coli genome with synthetic DNA; CGS minimizes crossovers1,4 between the synthetic DNA and the genome such that the output for each 100 kb replacement provides, without sequencing, the input for the next 100 kb replacement. Using CGS, we synthesized a 0.5 Mb section of the E. coli genome-a key intermediate in its total synthesis1-from five episomes in 10 days. By parallelizing CGS and combining it with rapid oligonucleotide synthesis and episome assembly5,6, along with rapid methods for compiling a single genome from strains bearing distinct synthetic genome sections1,7,8, we anticipate that it will be possible to synthesize entire E. coli genomes from functional designs in less than 2 months.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
J.W.C. is the founder of Constructive Bio. The MRC has filed a patent application covering the work described.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

