Genetic Engineering of Resident Bacteria in the Gut Microbiome
- PMID: 37382533
- PMCID: PMC10367592
- DOI: 10.1128/jb.00127-23
Genetic Engineering of Resident Bacteria in the Gut Microbiome
Abstract
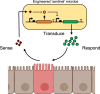
Techniques by which to genetically manipulate members of the microbiota enable both the evaluation of host-microbe interactions and an avenue by which to monitor and modulate human physiology. Genetic engineering applications have traditionally focused on model gut residents, such as Escherichia coli and lactic acid bacteria. However, emerging efforts by which to develop synthetic biology toolsets for "nonmodel" resident gut microbes could provide an improved foundation for microbiome engineering. As genome engineering tools come online, so too have novel applications for engineered gut microbes. Engineered resident gut bacteria facilitate investigations of the roles of microbes and their metabolites on host health and allow for potential live microbial biotherapeutics. Due to the rapid pace of discovery in this burgeoning field, this minireview highlights advancements in the genetic engineering of all resident gut microbes.
Keywords: CRISPR-Cas; metabolic engineering; microbiome; nonmodel organisms; synthetic biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, Casero D, Courtney H, Gonzalez A, Graeber TG, Hall AB, Lake K, Landers CJ, Mallick H, Plichta DR, Prasad M, Rahnavard G, Sauk J, Shungin D, Vázquez-Baeza Y, White RA, Braun J, Denson LA, Jansson JK, Knight R, Kugathasan S, McGovern DPB, Petrosino JF, Stappenbeck TS, Winter HS, Clish CB, Franzosa EA, Vlamakis H, Xavier RJ, Huttenhower C, IBDMDB Investigators . 2019. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569:655–662. doi: 10.1038/s41586-019-1237-9. - DOI - PMC - PubMed
-
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto J-M, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, et al. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60. doi: 10.1038/nature11450. - DOI - PubMed
-
- Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, Beghini F, Manara S, Karcher N, Pozzi C, Gandini S, Serrano D, Tarallo S, Francavilla A, Gallo G, Trompetto M, Ferrero G, Mizutani S, Shiroma H, Shiba S, Shibata T, Yachida S, Yamada T, Wirbel J, Schrotz-King P, Ulrich CM, Brenner H, Arumugam M, Bork P, Zeller G, Cordero F, Dias-Neto E, Setubal JC, Tett A, Pardini B, Rescigno M, Waldron L, Naccarati A, Segata N. 2019. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med 25:667–678. doi: 10.1038/s41591-019-0405-7. - DOI - PMC - PubMed
-
- Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, Fleck JS, Voigt AY, Palleja A, Ponnudurai R, Sunagawa S, Coelho LP, Schrotz-King P, Vogtmann E, Habermann N, Niméus E, Thomas AM, Manghi P, Gandini S, Serrano D, Mizutani S, Shiroma H, Shiba S, Shibata T, Yachida S, Yamada T, Waldron L, Naccarati A, Segata N, Sinha R, Ulrich CM, Brenner H, Arumugam M, Bork P, Zeller G. 2019. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med 25:679–689. doi: 10.1038/s41591-019-0406-6. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

