Promiscuous recognition of MR1 drives self-reactive mucosal-associated invariant T cell responses
- PMID: 37382893
- PMCID: PMC10309188
- DOI: 10.1084/jem.20221939
Promiscuous recognition of MR1 drives self-reactive mucosal-associated invariant T cell responses
Abstract
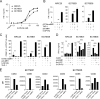
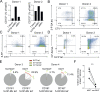
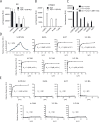
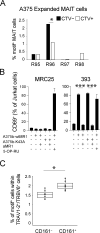
Mucosal-associated invariant T (MAIT) cells use canonical semi-invariant T cell receptors (TCR) to recognize microbial riboflavin precursors displayed by the antigen-presenting molecule MR1. The extent of MAIT TCR crossreactivity toward physiological, microbially unrelated antigens remains underexplored. We describe MAIT TCRs endowed with MR1-dependent reactivity to tumor and healthy cells in the absence of microbial metabolites. MAIT cells bearing TCRs crossreactive toward self are rare but commonly found within healthy donors and display T-helper-like functions in vitro. Experiments with MR1-tetramers loaded with distinct ligands revealed significant crossreactivity among MAIT TCRs both ex vivo and upon in vitro expansion. A canonical MAIT TCR was selected on the basis of extremely promiscuous MR1 recognition. Structural and molecular dynamic analyses associated promiscuity to unique TCRβ-chain features that were enriched within self-reactive MAIT cells of healthy individuals. Thus, self-reactive recognition of MR1 represents a functionally relevant indication of MAIT TCR crossreactivity, suggesting a potentially broader role of MAIT cells in immune homeostasis and diseases, beyond microbial immunosurveillance.
© 2023 Chancellor et al.
Conflict of interest statement
Disclosures: R. Simmons, V. Karuppiah, R. Suckling, M. Hock, and M. Lepore reported personal fees from Immunocore Ltd. during the conduct of the study and personal fees from Immunocore Ltd. outside the submitted work. No other disclosures were reported.
Figures







Similar articles
-
Mouse mucosal-associated invariant T cell receptor recognition of MR1 presenting the vitamin B metabolite, 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil.J Biol Chem. 2024 May;300(5):107229. doi: 10.1016/j.jbc.2024.107229. Epub 2024 Mar 25. J Biol Chem. 2024. PMID: 38537698 Free PMC article.
-
A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells.J Exp Med. 2014 Jul 28;211(8):1585-600. doi: 10.1084/jem.20140484. Epub 2014 Jul 21. J Exp Med. 2014. PMID: 25049336 Free PMC article.
-
MAIT cell-MR1 reactivity is highly conserved across multiple divergent species.J Biol Chem. 2024 Jun;300(6):107338. doi: 10.1016/j.jbc.2024.107338. Epub 2024 May 3. J Biol Chem. 2024. PMID: 38705391 Free PMC article.
-
Antigen Recognition by MR1-Reactive T Cells; MAIT Cells, Metabolites, and Remaining Mysteries.Front Immunol. 2020 Aug 27;11:1961. doi: 10.3389/fimmu.2020.01961. eCollection 2020. Front Immunol. 2020. PMID: 32973800 Free PMC article. Review.
-
MAIT, MR1, microbes and riboflavin: a paradigm for the co-evolution of invariant TCRs and restricting MHCI-like molecules?Immunogenetics. 2016 Aug;68(8):537-48. doi: 10.1007/s00251-016-0927-9. Epub 2016 Jul 8. Immunogenetics. 2016. PMID: 27393664 Review.
Cited by
-
Conserved allomorphs of MR1 drive the specificity of MR1-restricted TCRs.Front Oncol. 2024 Oct 3;14:1419528. doi: 10.3389/fonc.2024.1419528. eCollection 2024. Front Oncol. 2024. PMID: 39445059 Free PMC article.
-
Defenders or defectors: mucosal-associated invariant T cells in autoimmune diseases.Curr Opin Immunol. 2025 Apr;93:102542. doi: 10.1016/j.coi.2025.102542. Epub 2025 Feb 27. Curr Opin Immunol. 2025. PMID: 40020256 Review.
-
MR1 presents vitamin B6-related compounds for recognition by MR1-reactive T cells.Proc Natl Acad Sci U S A. 2024 Dec 3;121(49):e2414792121. doi: 10.1073/pnas.2414792121. Epub 2024 Nov 26. Proc Natl Acad Sci U S A. 2024. PMID: 39589872 Free PMC article.
-
Regulators and Conductors of Immunity: Natural Immune System in Health and Autoimmunity.Int J Mol Sci. 2025 Jun 5;26(11):5413. doi: 10.3390/ijms26115413. Int J Mol Sci. 2025. PMID: 40508221 Free PMC article. Review.
-
Molecular Insights Into MR1-Mediated T Cell Immunity: Lessons Learned and Unanswered Questions.Immunol Rev. 2025 May;331(1):e70033. doi: 10.1111/imr.70033. Immunol Rev. 2025. PMID: 40338831 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

