Methanol-based biomanufacturing of fuels and chemicals using native and synthetic methylotrophs
- PMID: 37384124
- PMCID: PMC10293595
- DOI: 10.1016/j.synbio.2023.06.001
Methanol-based biomanufacturing of fuels and chemicals using native and synthetic methylotrophs
Abstract
Methanol has recently gained significant attention as a potential carbon substrate for the production of fuels and chemicals, owing to its high degree of reduction, abundance, and low price. Native methylotrophic yeasts and bacteria have been investigated for the production of fuels and chemicals. Alternatively, synthetic methylotrophic strains are also being developed by reconstructing methanol utilization pathways in model microorganisms, such as Escherichia coli. Owing to the complex metabolic pathways, limited availability of genetic tools, and methanol/formaldehyde toxicity, the high-level production of target products for industrial applications are still under development to satisfy commercial feasibility. This article reviews the production of biofuels and chemicals by native and synthetic methylotrophic microorganisms. It also highlights the advantages and limitations of both types of methylotrophs and provides an overview of ways to improve their efficiency for the production of fuels and chemicals from methanol.
Keywords: Bacillus methanolicus; Escherichia coli; Methylobacterium extorquens; Native methylotrophs; Pichia pastoris; Synthetic methylotrophs.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


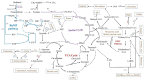
 H4F, N5,N10-methylenetetrahydromethanopterin; H4MPT, Tetrahydromethanopterin; H4F, tetrahydrofolate; PEP, Phosphoenolpyruvate; H6P, Hexulose-6-phosphate; F6P, Fructose-6-phosphate; FBP, Fructose 1,6-bisphophate; DHAP, Dihydroxyacetone phosphate; GAP, Glyceraldehyde 3-phosphate; Xu5P, Xylulose 5-phosphate; E4P, Erythrose 4-phosphate; S7P, sedoheptulose-7-phosphate; R5P, ribose-5-phosphate; Ru5P, Ribulose-5-phosphate; CO2, Carbon dioxide; 3-HP, 3- Hydroxy propionic acid; PHB, Poly (3-Hydroxybutyrate); 2-HIBA, 2-Hydroxyisobutyric acid; FPP, Farnesyl diphosphate; P (3HB-co-3HV-co-3HHX), Poly (3-hydroxybutyrate-co-3-hydroxyvalerate-co-3-hydroxyhexanoate); GABA, γ-aminobutyric acid.
H4F, N5,N10-methylenetetrahydromethanopterin; H4MPT, Tetrahydromethanopterin; H4F, tetrahydrofolate; PEP, Phosphoenolpyruvate; H6P, Hexulose-6-phosphate; F6P, Fructose-6-phosphate; FBP, Fructose 1,6-bisphophate; DHAP, Dihydroxyacetone phosphate; GAP, Glyceraldehyde 3-phosphate; Xu5P, Xylulose 5-phosphate; E4P, Erythrose 4-phosphate; S7P, sedoheptulose-7-phosphate; R5P, ribose-5-phosphate; Ru5P, Ribulose-5-phosphate; CO2, Carbon dioxide; 3-HP, 3- Hydroxy propionic acid; PHB, Poly (3-Hydroxybutyrate); 2-HIBA, 2-Hydroxyisobutyric acid; FPP, Farnesyl diphosphate; P (3HB-co-3HV-co-3HHX), Poly (3-hydroxybutyrate-co-3-hydroxyvalerate-co-3-hydroxyhexanoate); GABA, γ-aminobutyric acid.
 H4F, N5,N10-methylenetetrahydromethanopterin; CO2, Carbon dioxide; H6P, Hexulose-6-phosphate; F6P, Fructose-6-phosphate; FBP, Fructose 1,6-bisphosphate; DHAP, Dihydroxyacetone phosphate; GAP, Glyceraldehyde 3-phosphate; Ru5P, Ribulose 5-Phosphate; G6P, glucose-6-phosphate; 6-PG, 6-phosphogluconate; G3P, glyceraldehyde-3-phosphate; 2-PG, 2-phosphoglycerate; H4MPT, Tetrahydromethanopterin; H4F, tetrahydrofolate; Xu5P, Xylulose 5-phosphate; Gly3P, glycerol 3-phosphate; ADP, Adenosine diphosphate; ATP, Adenosine triphosphate, NAD(P)+, Nicotinamide adenine dinucleotide phosphate; NAD(P)H, reduced form of NAD(P)+; NAD+, Nicotinamide adenine dinucleotide; NADH, Reduced form of NAD+; 3PGA, 3-phosphoglycerate; RuBP, Ribulose 1,5-bisphosphate.
H4F, N5,N10-methylenetetrahydromethanopterin; CO2, Carbon dioxide; H6P, Hexulose-6-phosphate; F6P, Fructose-6-phosphate; FBP, Fructose 1,6-bisphosphate; DHAP, Dihydroxyacetone phosphate; GAP, Glyceraldehyde 3-phosphate; Ru5P, Ribulose 5-Phosphate; G6P, glucose-6-phosphate; 6-PG, 6-phosphogluconate; G3P, glyceraldehyde-3-phosphate; 2-PG, 2-phosphoglycerate; H4MPT, Tetrahydromethanopterin; H4F, tetrahydrofolate; Xu5P, Xylulose 5-phosphate; Gly3P, glycerol 3-phosphate; ADP, Adenosine diphosphate; ATP, Adenosine triphosphate, NAD(P)+, Nicotinamide adenine dinucleotide phosphate; NAD(P)H, reduced form of NAD(P)+; NAD+, Nicotinamide adenine dinucleotide; NADH, Reduced form of NAD+; 3PGA, 3-phosphoglycerate; RuBP, Ribulose 1,5-bisphosphate.References
-
- Ali A., Shen P.K. Recent advances in graphene-based platinum and palladium electrocatalysts for the methanol oxidation reaction. J Mater Chem. 2019;7(39):22189–22217. doi: 10.1039/c9ta06088j. - DOI
-
- Khirsariya P., Mewada R.K. Single step oxidation of methane to methanol–towards better understanding. Procedia Eng. 2013;51:409–415. doi: 10.1016/j.proeng.2013.01.057. - DOI
LinkOut - more resources
Full Text Sources

