Analytical validation of quantitative SARS-CoV-2 subgenomic and viral load laboratory developed tests conducted on the Panther Fusion® (Hologic) with preliminary application to clinical samples
- PMID: 37384714
- PMCID: PMC10309597
- DOI: 10.1371/journal.pone.0287576
Analytical validation of quantitative SARS-CoV-2 subgenomic and viral load laboratory developed tests conducted on the Panther Fusion® (Hologic) with preliminary application to clinical samples
Abstract
Objective: Validate the performance characteristics of two analyte specific, laboratory developed tests (LDTs) for the quantification of SARS-CoV-2 subgenomic RNA (sgRNA) and viral load on the Hologic Panther Fusion® using the Open Access functionality.
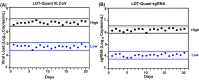
Methods: Custom-designed primers/probe sets targeting the SARS-CoV-2 Envelope gene (E) and subgenomic E were optimized. A 20-day performance validation following laboratory developed test requirements was conducted to assess assay precision, accuracy, analytical sensitivity/specificity, lower limit of detection and reportable range.
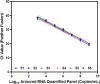
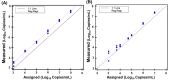
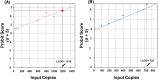
Results: Quantitative SARS-CoV-2 sgRNA (LDT-Quant sgRNA) assay, which measures intermediates of replication, and viral load (LDT-Quant VLCoV) assay demonstrated acceptable performance. Both assays were linear with an R2 and slope equal to 0.99 and 1.00, respectively. Assay precision was evaluated between 4-6 Log10 with a maximum CV of 2.6% and 2.5% for LDT-Quant sgRNA and LDT-Quant VLCoV respectively. Using negative or positive SARS-CoV-2 human nasopharyngeal swab samples, both assays were accurate (kappa coefficient of 1.00 and 0.92). Common respiratory flora and other viral pathogens were not detected and did not interfere with the detection or quantification by either assay. Based on 95% detection, the assay LLODs were 729 and 1206 Copies/mL for the sgRNA and VL load LDTs, respectively.
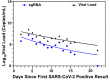
Conclusion: The LDT-Quant sgRNA and LDT-Quant VLCoV demonstrated good analytical performance. These assays could be further investigated as alternative monitoring assays for viral replication; and thus, medical management in clinical settings which could inform isolation/quarantine requirements.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Lefeuvre C, Pivert A, Przyrowski E, Bouthry E, Darviot E, Mahieu R, et al.. Comparison of performance between three SARS-CoV-2 molecular assays (Aptima, Laboratory Developed Test-Fusion, and R-GENE(R)) with special attention to turnaround time, a key point in laboratory management. J Med Virol. 2022. Epub 2022/02/26. doi: 10.1002/jmv.27675 . - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

