Dissecting the roles of MBD2 isoforms and domains in regulating NuRD complex function during cellular differentiation
- PMID: 37385984
- PMCID: PMC10310694
- DOI: 10.1038/s41467-023-39551-w
Dissecting the roles of MBD2 isoforms and domains in regulating NuRD complex function during cellular differentiation
Abstract
The Nucleosome Remodeling and Deacetylation (NuRD) complex is a crucial regulator of cellular differentiation. Two members of the Methyl-CpG-binding domain (MBD) protein family, MBD2 and MBD3, are known to be integral, but mutually exclusive subunits of the NuRD complex. Several MBD2 and MBD3 isoforms are present in mammalian cells, resulting in distinct MBD-NuRD complexes. Whether these different complexes serve distinct functional activities during differentiation is not fully explored. Based on the essential role of MBD3 in lineage commitment, we systematically investigated a diverse set of MBD2 and MBD3 variants for their potential to rescue the differentiation block observed for mouse embryonic stem cells (ESCs) lacking MBD3. While MBD3 is indeed crucial for ESC differentiation to neuronal cells, it functions independently of its MBD domain. We further identify that MBD2 isoforms can replace MBD3 during lineage commitment, however with different potential. Full-length MBD2a only partially rescues the differentiation block, while MBD2b, an isoform lacking an N-terminal GR-rich repeat, fully rescues the Mbd3 KO phenotype. In case of MBD2a, we further show that removing the methylated DNA binding capacity or the GR-rich repeat enables full redundancy to MBD3, highlighting the synergistic requirements for these domains in diversifying NuRD complex function.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



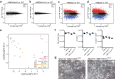
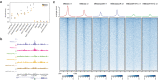
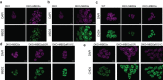
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

