Down-regulation of peptidylarginine deiminase type 1 in reconstructed human epidermis disturbs nucleophagy in the granular layer and affects barrier function
- PMID: 37385992
- PMCID: PMC10310762
- DOI: 10.1038/s41420-023-01509-8
Down-regulation of peptidylarginine deiminase type 1 in reconstructed human epidermis disturbs nucleophagy in the granular layer and affects barrier function
Abstract
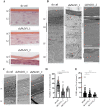
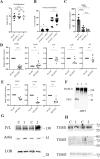
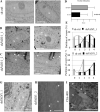
Deimination is a post-translational modification catalyzed by a family of enzymes named peptidylarginine deiminases (PADs). PADs transform arginine residues of protein substrates into citrulline. Deimination has been associated with numerous physiological and pathological processes. In human skin, three PADs are expressed (PAD1-3). While PAD3 is important for hair shape formation, the role of PAD1 is less clear. To decipher the main role(s) of PAD1 in epidermal differentiation, its expression was down-regulated using lentivirus-mediated shRNA interference in primary keratinocytes and in three-dimensional reconstructed human epidermis (RHE). Compared to normal RHEs, down-regulation of PAD1 caused a drastic reduction in deiminated proteins. Whereas proliferation of keratinocytes was not affected, their differentiation was disturbed at molecular, cellular and functional levels. The number of corneocyte layers was significantly reduced, expression of filaggrin and cornified cell envelope components, such as loricrin and transglutaminases, was down-regulated, epidermal permeability increased and trans-epidermal-electric resistance diminished drastically. Keratohyalin granule density decreased and nucleophagy in the granular layer was disturbed. These results demonstrate that PAD1 is the main regulator of protein deimination in RHE. Its deficiency alters epidermal homeostasis, affecting the differentiation of keratinocytes, especially the cornification process, a special kind of programmed cell death.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Figures

References
-
- Chavanas S, Méchin M-C, Takahara H, Kawada A, Nachat R, Serre G, et al. Comparative analysis of the mouse and human peptidylarginine deiminase gene clusters reveals highly conserved non-coding segments and a new human gene, PADI6. Gene. 2004;330:19–27. doi: 10.1016/j.gene.2003.12.038. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

