The proteomic landscape of soft tissue sarcomas
- PMID: 37386008
- PMCID: PMC10310735
- DOI: 10.1038/s41467-023-39486-2
The proteomic landscape of soft tissue sarcomas
Abstract
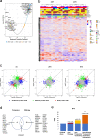
Soft tissue sarcomas (STS) are rare and diverse mesenchymal cancers with limited treatment options. Here we undertake comprehensive proteomic profiling of tumour specimens from 321 STS patients representing 11 histological subtypes. Within leiomyosarcomas, we identify three proteomic subtypes with distinct myogenesis and immune features, anatomical site distribution and survival outcomes. Characterisation of undifferentiated pleomorphic sarcomas and dedifferentiated liposarcomas with low infiltrating CD3 + T-lymphocyte levels nominates the complement cascade as a candidate immunotherapeutic target. Comparative analysis of proteomic and transcriptomic profiles highlights the proteomic-specific features for optimal risk stratification in angiosarcomas. Finally, we define functional signatures termed Sarcoma Proteomic Modules which transcend histological subtype classification and show that a vesicle transport protein signature is an independent prognostic factor for distant metastasis. Our study highlights the utility of proteomics for identifying molecular subgroups with implications for risk stratification and therapy selection and provides a rich resource for future sarcoma research.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
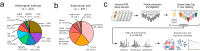




References
-
- WHO Classification of Tumours Editorial Board. Soft Tissue and Bone Tumours (International Agency for Research on Cancer, 2020).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

