Single-cell transcriptomics uncovers EGFR signaling-mediated gastric progenitor cell differentiation in stomach homeostasis
- PMID: 37386010
- PMCID: PMC10310803
- DOI: 10.1038/s41467-023-39113-0
Single-cell transcriptomics uncovers EGFR signaling-mediated gastric progenitor cell differentiation in stomach homeostasis
Erratum in
-
Author Correction: Single-cell transcriptomics uncovers EGFR signaling-mediated gastric progenitor cell differentiation in stomach homeostasis.Nat Commun. 2023 Oct 17;14(1):6540. doi: 10.1038/s41467-023-41823-4. Nat Commun. 2023. PMID: 37848419 Free PMC article. No abstract available.
Abstract
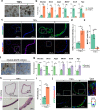
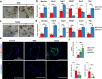
Defects in gastric progenitor cell differentiation are associated with various gastric disorders, including atrophic gastritis, intestinal metaplasia, and gastric cancer. However, the mechanisms underlying the multilineage differentiation of gastric progenitor cells during healthy homeostasis remain poorly understood. Here, using a single-cell RNA sequencing method, Quartz-Seq2, we analyzed the gene expression dynamics of progenitor cell differentiation toward pit cell, neck cell, and parietal cell lineages in healthy adult mouse corpus tissues. Enrichment analysis of pseudotime-dependent genes and a gastric organoid assay revealed that EGFR-ERK signaling promotes pit cell differentiation, whereas NF-κB signaling maintains gastric progenitor cells in an undifferentiated state. In addition, pharmacological inhibition of EGFR in vivo resulted in a decreased number of pit cells. Although activation of EGFR signaling in gastric progenitor cells has been suggested as one of the major inducers of gastric cancers, our findings unexpectedly identified that EGFR signaling exerts a differentiation-promoting function, not a mitogenic function, in normal gastric homeostasis.
© 2023. The Author(s).
Conflict of interest statement
Y.S. and I.N. consult for Knowledge Palette, Inc. and are on their Scientific Advisory Board. The other authors declare no competing interests.
Figures





References
-
- Neuburger P, Lewin M, de Recherche C, Bonfils S. Parietal and chief cell populations in four cases of the Zollinger-Ellison syndrome. Gastroenterology. 1972;63:937–942. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

