Single cell Hi-C identifies plastic chromosome conformations underlying the gastrulation enhancer landscape
- PMID: 37386027
- PMCID: PMC10310791
- DOI: 10.1038/s41467-023-39549-4
Single cell Hi-C identifies plastic chromosome conformations underlying the gastrulation enhancer landscape
Abstract
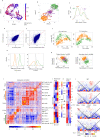
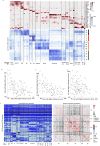
Embryonic development involves massive proliferation and differentiation of cell lineages. This must be supported by chromosome replication and epigenetic reprogramming, but how proliferation and cell fate acquisition are balanced in this process is not well understood. Here we use single cell Hi-C to map chromosomal conformations in post-gastrulation mouse embryo cells and study their distributions and correlations with matching embryonic transcriptional atlases. We find that embryonic chromosomes show a remarkably strong cell cycle signature. Despite that, replication timing, chromosome compartment structure, topological associated domains (TADs) and promoter-enhancer contacts are shown to be variable between distinct epigenetic states. About 10% of the nuclei are identified as primitive erythrocytes, showing exceptionally compact and organized compartment structure. The remaining cells are broadly associated with ectoderm and mesoderm identities, showing only mild differentiation of TADs and compartment structures, but more specific localized contacts in hundreds of ectoderm and mesoderm promoter-enhancer pairs. The data suggest that while fully committed embryonic lineages can rapidly acquire specific chromosomal conformations, most embryonic cells are showing plastic signatures driven by complex and intermixed enhancer landscapes.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

