The mRNA methyltransferase Mettl3 modulates cytokine mRNA stability and limits functional responses in mast cells
- PMID: 37386028
- PMCID: PMC10310798
- DOI: 10.1038/s41467-023-39614-y
The mRNA methyltransferase Mettl3 modulates cytokine mRNA stability and limits functional responses in mast cells
Abstract
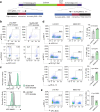
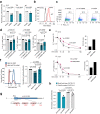
Mast cells are central players in allergy and asthma, and their dysregulated responses lead to reduced quality of life and life-threatening conditions such as anaphylaxis. The RNA modification N6-methyladenosine (m6A) has a prominent impact on immune cell functions, but its role in mast cells remains unexplored. Here, by optimizing tools to genetically manipulate primary mast cells, we reveal that the m6A mRNA methyltransferase complex modulates mast cell proliferation and survival. Depletion of the catalytic component Mettl3 exacerbates effector functions in response to IgE and antigen complexes, both in vitro and in vivo. Mechanistically, deletion of Mettl3 or Mettl14, another component of the methyltransferase complex, lead to the enhanced expression of inflammatory cytokines. By focusing on one of the most affected mRNAs, namely the one encoding the cytokine IL-13, we find that it is methylated in activated mast cells, and that Mettl3 affects its transcript stability in an enzymatic activity-dependent manner, requiring consensus m6A sites in the Il13 3'-untranslated region. Overall, we reveal that the m6A machinery is essential in mast cells to sustain growth and to restrain inflammatory responses.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

