Parabacteroides distasonis uses dietary inulin to suppress NASH via its metabolite pentadecanoic acid
- PMID: 37386075
- PMCID: PMC10390331
- DOI: 10.1038/s41564-023-01418-7
Parabacteroides distasonis uses dietary inulin to suppress NASH via its metabolite pentadecanoic acid
Abstract
Non-alcoholic steatohepatitis (NASH) is the severe form of non-alcoholic fatty liver disease, and is characterized by liver inflammation and fat accumulation. Dietary interventions, such as fibre, have been shown to alleviate this metabolic disorder in mice via the gut microbiota. Here, we investigated the mechanistic role of the gut microbiota in ameliorating NASH via dietary fibre in mice. Soluble fibre inulin was found to be more effective than insoluble fibre cellulose to suppress NASH progression in mice, as shown by reduced hepatic steatosis, necro-inflammation, ballooning and fibrosis. We employed stable isotope probing to trace the incorporation of 13C-inulin into gut bacterial genomes and metabolites during NASH progression. Shotgun metagenome sequencing revealed that the commensal Parabacteroides distasonis was enriched by 13C-inulin. Integration of 13C-inulin metagenomes and metabolomes suggested that P. distasonis used inulin to produce pentadecanoic acid, an odd-chain fatty acid, which was confirmed in vitro and in germ-free mice. P. distasonis or pentadecanoic acid was protective against NASH in mice. Mechanistically, inulin, P. distasonis or pentadecanoic acid restored gut barrier function in NASH models, which reduced serum lipopolysaccharide and liver pro-inflammatory cytokine expression. Overall this shows that gut microbiota members can use dietary fibre to generate beneficial metabolites to suppress metabolic disease.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

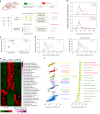
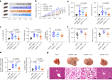





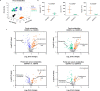

Comment in
-
Tracking microbiota fibre breakdown to treat NASH.Nat Microbiol. 2023 Aug;8(8):1382-1383. doi: 10.1038/s41564-023-01440-9. Nat Microbiol. 2023. PMID: 37488258 No abstract available.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

