mTORC2-NDRG1-CDC42 axis couples fasting to mitochondrial fission
- PMID: 37386153
- PMCID: PMC10344787
- DOI: 10.1038/s41556-023-01163-3
mTORC2-NDRG1-CDC42 axis couples fasting to mitochondrial fission
Abstract
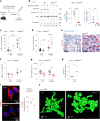
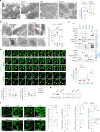
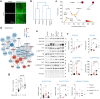
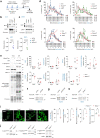
Fasting triggers diverse physiological adaptations including increases in circulating fatty acids and mitochondrial respiration to facilitate organismal survival. The mechanisms driving mitochondrial adaptations and respiratory sufficiency during fasting remain incompletely understood. Here we show that fasting or lipid availability stimulates mTORC2 activity. Activation of mTORC2 and phosphorylation of its downstream target NDRG1 at serine 336 sustains mitochondrial fission and respiratory sufficiency. Time-lapse imaging shows that NDRG1, but not the phosphorylation-deficient NDRG1Ser336Ala mutant, engages with mitochondria to facilitate fission in control cells, as well as in those lacking DRP1. Using proteomics, a small interfering RNA screen, and epistasis experiments, we show that mTORC2-phosphorylated NDRG1 cooperates with small GTPase CDC42 and effectors and regulators of CDC42 to orchestrate fission. Accordingly, RictorKO, NDRG1Ser336Ala mutants and Cdc42-deficient cells each display mitochondrial phenotypes reminiscent of fission failure. During nutrient surplus, mTOR complexes perform anabolic functions; however, paradoxical reactivation of mTORC2 during fasting unexpectedly drives mitochondrial fission and respiration.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures












References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

