MultiVI: deep generative model for the integration of multimodal data
- PMID: 37386189
- PMCID: PMC10406609
- DOI: 10.1038/s41592-023-01909-9
MultiVI: deep generative model for the integration of multimodal data
Abstract
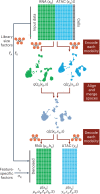
Jointly profiling the transcriptome, chromatin accessibility and other molecular properties of single cells offers a powerful way to study cellular diversity. Here we present MultiVI, a probabilistic model to analyze such multiomic data and leverage it to enhance single-modality datasets. MultiVI creates a joint representation that allows an analysis of all modalities included in the multiomic input data, even for cells for which one or more modalities are missing. It is available at scvi-tools.org .
© 2023. The Author(s).
Conflict of interest statement
N.Y. is an adviser and/or has equity in Cellarity, Celsius Therapeutics and Rheos Medicines. T.A is an employee of Vevo Therapeutics. All other authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

