SpatialSort: a Bayesian model for clustering and cell population annotation of spatial proteomics data
- PMID: 37387130
- PMCID: PMC10311307
- DOI: 10.1093/bioinformatics/btad242
SpatialSort: a Bayesian model for clustering and cell population annotation of spatial proteomics data
Abstract
Motivation: Recent advances in spatial proteomics technologies have enabled the profiling of dozens of proteins in thousands of single cells in situ. This has created the opportunity to move beyond quantifying the composition of cell types in tissue, and instead probe the spatial relationships between cells. However, most current methods for clustering data from these assays only consider the expression values of cells and ignore the spatial context. Furthermore, existing approaches do not account for prior information about the expected cell populations in a sample.
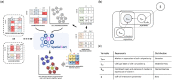
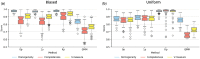
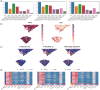
Results: To address these shortcomings, we developed SpatialSort, a spatially aware Bayesian clustering approach that allows for the incorporation of prior biological knowledge. Our method is able to account for the affinities of cells of different types to neighbour in space, and by incorporating prior information about expected cell populations, it is able to simultaneously improve clustering accuracy and perform automated annotation of clusters. Using synthetic and real data, we show that by using spatial and prior information SpatialSort improves clustering accuracy. We also demonstrate how SpatialSort can perform label transfer between spatial and nonspatial modalities through the analysis of a real world diffuse large B-cell lymphoma dataset.
Availability and implementation: Source code is available on Github at: https://github.com/Roth-Lab/SpatialSort.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
C.H. and A.G. are employees of Bristol Myers Squibb. The other authors declare that they have no competing interests.
Figures


References
-
- Ali HR, Jackson HW, Zanotelli VRT. et al. ; CRUK IMAXT Grand Challenge Team. Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer. Nat Cancer 2020;1:163–75. - PubMed
-
- Bishop CM. Pattern Recognition and Machine Learning. Berlin, Heidelberg: Springer-Verlag; 2006.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

