Phylogenomic branch length estimation using quartets
- PMID: 37387151
- PMCID: PMC10311336
- DOI: 10.1093/bioinformatics/btad221
Phylogenomic branch length estimation using quartets
Abstract
Motivation: Branch lengths and topology of a species tree are essential in most downstream analyses, including estimation of diversification dates, characterization of selection, understanding adaptation, and comparative genomics. Modern phylogenomic analyses often use methods that account for the heterogeneity of evolutionary histories across the genome due to processes such as incomplete lineage sorting. However, these methods typically do not generate branch lengths in units that are usable by downstream applications, forcing phylogenomic analyses to resort to alternative shortcuts such as estimating branch lengths by concatenating gene alignments into a supermatrix. Yet, concatenation and other available approaches for estimating branch lengths fail to address heterogeneity across the genome.
Results: In this article, we derive expected values of gene tree branch lengths in substitution units under an extension of the multispecies coalescent (MSC) model that allows substitutions with varying rates across the species tree. We present CASTLES, a new technique for estimating branch lengths on the species tree from estimated gene trees that uses these expected values, and our study shows that CASTLES improves on the most accurate prior methods with respect to both speed and accuracy.
Availability and implementation: CASTLES is available at https://github.com/ytabatabaee/CASTLES.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures
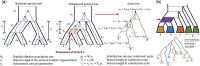
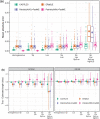
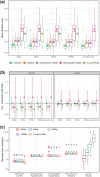
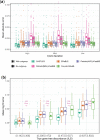
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

