A gated graph transformer for protein complex structure quality assessment and its performance in CASP15
- PMID: 37387159
- PMCID: PMC10311325
- DOI: 10.1093/bioinformatics/btad203
A gated graph transformer for protein complex structure quality assessment and its performance in CASP15
Abstract
Motivation: Proteins interact to form complexes to carry out essential biological functions. Computational methods such as AlphaFold-multimer have been developed to predict the quaternary structures of protein complexes. An important yet largely unsolved challenge in protein complex structure prediction is to accurately estimate the quality of predicted protein complex structures without any knowledge of the corresponding native structures. Such estimations can then be used to select high-quality predicted complex structures to facilitate biomedical research such as protein function analysis and drug discovery.
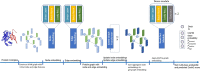
Results: In this work, we introduce a new gated neighborhood-modulating graph transformer to predict the quality of 3D protein complex structures. It incorporates node and edge gates within a graph transformer framework to control information flow during graph message passing. We trained, evaluated and tested the method (called DProQA) on newly-curated protein complex datasets before the 15th Critical Assessment of Techniques for Protein Structure Prediction (CASP15) and then blindly tested it in the 2022 CASP15 experiment. The method was ranked 3rd among the single-model quality assessment methods in CASP15 in terms of the ranking loss of TM-score on 36 complex targets. The rigorous internal and external experiments demonstrate that DProQA is effective in ranking protein complex structures.
Availability and implementation: The source code, data, and pre-trained models are available at https://github.com/jianlin-cheng/DProQA.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures



References
-
- Athanasios A, Charalampos V, Vasileios T. et al. Protein–protein interaction (PPI) network: recent advances in drug discovery. Curr Drug Metab 2017;18:5–10. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

