Performance and accuracy evaluation of reference panels for genotype imputation in sub-Saharan African populations
- PMID: 37388906
- PMCID: PMC10300601
- DOI: 10.1016/j.xgen.2023.100332
Performance and accuracy evaluation of reference panels for genotype imputation in sub-Saharan African populations
Abstract
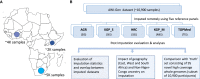
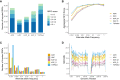
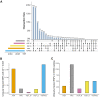
Based on evaluations of imputation performed on a genotype dataset consisting of about 11,000 sub-Saharan African (SSA) participants, we show Trans-Omics for Precision Medicine (TOPMed) and the African Genome Resource (AGR) to be currently the best panels for imputing SSA datasets. We report notable differences in the number of single-nucleotide polymorphisms (SNPs) that are imputed by different panels in datasets from East, West, and South Africa. Comparisons with a subset of 95 SSA high-coverage whole-genome sequences (WGSs) show that despite being about 20-fold smaller, the AGR imputed dataset has higher concordance with the WGSs. Moreover, the level of concordance between imputed and WGS datasets was strongly influenced by the extent of Khoe-San ancestry in a genome, highlighting the need for integration of not only geographically but also ancestrally diverse WGS data in reference panels for further improvement in imputation of SSA datasets. Approaches that integrate imputed data from different panels could also lead to better imputation.
Keywords: AGR; Africa; GWAS; TOPMed; imputation; imputation accuracy; non-reference discordance rate; reference panel; whole-genome sequence.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Das S., Abecasis G.R., Browning B.L. Genotype imputation from large reference panels. Annu. Rev. Genomics Hum. Genet. 2018;19:73–96. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

