Genome-wide structural variant analysis identifies risk loci for non-Alzheimer's dementias
- PMID: 37388914
- PMCID: PMC10300553
- DOI: 10.1016/j.xgen.2023.100316
Genome-wide structural variant analysis identifies risk loci for non-Alzheimer's dementias
Abstract
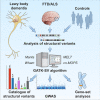
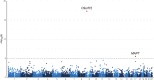
We characterized the role of structural variants, a largely unexplored type of genetic variation, in two non-Alzheimer's dementias, namely Lewy body dementia (LBD) and frontotemporal dementia (FTD)/amyotrophic lateral sclerosis (ALS). To do this, we applied an advanced structural variant calling pipeline (GATK-SV) to short-read whole-genome sequence data from 5,213 European-ancestry cases and 4,132 controls. We discovered, replicated, and validated a deletion in TPCN1 as a novel risk locus for LBD and detected the known structural variants at the C9orf72 and MAPT loci as associated with FTD/ALS. We also identified rare pathogenic structural variants in both LBD and FTD/ALS. Finally, we assembled a catalog of structural variants that can be mined for new insights into the pathogenesis of these understudied forms of dementia.
Keywords: Lewy body dementia; amyotrophic lateral sclerosis; case-control study; frontotemporal dementia; genome-wide association study; non–Alzheimer's dementia; resource; structural variant.
Conflict of interest statement
S.W.S. serves on the Scientific Advisory Council of the Lewy Body Dementia Association and the Multiple System Atrophy Coalition. S.W.S. and B.J.T. receive research support from Cerevel Therapeutics. B.J.T. holds patents on the clinical testing and therapeutic implications of the C9orf72 repeat expansion. H.R.M. is employed by the University College London. In the last 12 months, he reports paid consultancy from Roche and Amylyx; lecture fees/honoraria from BMJ, Kyowa Kirin, and Movement Disorders Society; and research grants from Parkinson’s UK, Cure Parkinson’s Trust, PSP Association, Medical Research Council, and the Michael J Fox Foundation. H.R.M. is a co-applicant on a patent application related to C9orf72 “method for diagnosing a neurodegenerative disease” (PCT/GB2012/052140).
Figures





References
-
- Blauwendraat C., Pletnikova O., Geiger J.T., Murphy N.A., Abramzon Y., Rudow G., Mamais A., Sabir M.S., Crain B., Ahmed S., et al. Genetic analysis of neurodegenerative diseases in a pathology cohort. Neurobiol. Aging. 2019;76 doi: 10.1016/j.neurobiolaging.2018.11.007. 214.e1–214214.e9. - DOI - PMC - PubMed
Grants and funding
- P30 AG072975/AG/NIA NIH HHS/United States
- K08 AG065463/AG/NIA NIH HHS/United States
- U24 AG021886/AG/NIA NIH HHS/United States
- U01 AG061356/AG/NIA NIH HHS/United States
- P30 AG072980/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- P50 NS038377/NS/NINDS NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- P30 AG066507/AG/NIA NIH HHS/United States
- R35 NS127253/NS/NINDS NIH HHS/United States
- T32 HG002295/HG/NHGRI NIH HHS/United States
- ZIA AG000935/ImNIH/Intramural NIH HHS/United States
- P30 AG066546/AG/NIA NIH HHS/United States
- RF1 AG057473/AG/NIA NIH HHS/United States
- ZIA NS003154/ImNIH/Intramural NIH HHS/United States
- U24 NS072026/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

