Antimicrobial resistance and mechanisms of epigenetic regulation
- PMID: 37389209
- PMCID: PMC10306973
- DOI: 10.3389/fcimb.2023.1199646
Antimicrobial resistance and mechanisms of epigenetic regulation
Abstract
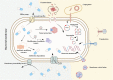
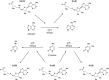
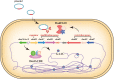
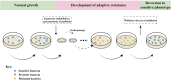
The rampant use of antibiotics in animal husbandry, farming and clinical disease treatment has led to a significant issue with pathogen resistance worldwide over the past decades. The classical mechanisms of resistance typically investigate antimicrobial resistance resulting from natural resistance, mutation, gene transfer and other processes. However, the emergence and development of bacterial resistance cannot be fully explained from a genetic and biochemical standpoint. Evolution necessitates phenotypic variation, selection, and inheritance. There are indications that epigenetic modifications also play a role in antimicrobial resistance. This review will specifically focus on the effects of DNA modification, histone modification, rRNA methylation and the regulation of non-coding RNAs expression on antimicrobial resistance. In particular, we highlight critical work that how DNA methyltransferases and non-coding RNAs act as transcriptional regulators that allow bacteria to rapidly adapt to environmental changes and control their gene expressions to resist antibiotic stress. Additionally, it will delve into how Nucleolar-associated proteins in bacteria perform histone functions akin to eukaryotes. Epigenetics, a non-classical regulatory mechanism of bacterial resistance, may offer new avenues for antibiotic target selection and the development of novel antibiotics.
Keywords: DNA modification; antimicrobial resistance; epigenetic drugs; epigenetics; non-coding RNAs; rRNA methylation.
Copyright © 2023 Wang, Yu and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
-
Directed evolution of the rRNA methylating enzyme Cfr reveals molecular basis of antibiotic resistance.Elife. 2022 Jan 11;11:e70017. doi: 10.7554/eLife.70017. Elife. 2022. PMID: 35015630 Free PMC article.
-
Epigenetic-Mediated Antimicrobial Resistance: Host versus Pathogen Epigenetic Alterations.Antibiotics (Basel). 2022 Jun 16;11(6):809. doi: 10.3390/antibiotics11060809. Antibiotics (Basel). 2022. PMID: 35740215 Free PMC article. Review.
-
Antibiotic Resistance and Epigenetics: More to It than Meets the Eye.Antimicrob Agents Chemother. 2020 Jan 27;64(2):e02225-19. doi: 10.1128/AAC.02225-19. Print 2020 Jan 27. Antimicrob Agents Chemother. 2020. PMID: 31740560 Free PMC article. Review.
-
An Update of Epigenetic Drugs for the Treatment of Cancers and Brain Diseases: A Comprehensive Review.Genes (Basel). 2023 Apr 6;14(4):873. doi: 10.3390/genes14040873. Genes (Basel). 2023. PMID: 37107631 Free PMC article. Review.
Cited by
-
The Emerging Role of IGF2BP2 in Cancer Therapy Resistance: From Molecular Mechanism to Future Potential.Int J Mol Sci. 2024 Nov 12;25(22):12150. doi: 10.3390/ijms252212150. Int J Mol Sci. 2024. PMID: 39596216 Free PMC article. Review.
-
Accurate bacterial outbreak tracing with Oxford Nanopore sequencing and reduction of methylation-induced errors.Genome Res. 2024 Nov 20;34(11):2039-2047. doi: 10.1101/gr.278848.123. Genome Res. 2024. PMID: 39443152 Free PMC article.
-
Prevalence and Genomic Characterization of Vibrio parahaemolyticus Isolated from a Vast Amount of Aquatic Products in Huzhou, China.Foods. 2025 Jul 15;14(14):2481. doi: 10.3390/foods14142481. Foods. 2025. PMID: 40724302 Free PMC article.
-
One Earth: The Equilibrium between the Human and the Bacterial Worlds.Int J Mol Sci. 2023 Oct 10;24(20):15047. doi: 10.3390/ijms242015047. Int J Mol Sci. 2023. PMID: 37894729 Free PMC article. Review.
-
Neutrophil extracellular traps in homeostasis and disease.Signal Transduct Target Ther. 2024 Sep 20;9(1):235. doi: 10.1038/s41392-024-01933-x. Signal Transduct Target Ther. 2024. PMID: 39300084 Free PMC article. Review.
References
-
- Ashok A. K., Gnanasekaran T. S., Santosh K. H. S., Srikanth K., Prakash N., Gollapalli P. (2023). High-throughput screening and molecular dynamics simulations of natural products targeting LuxS/AI-2 system as a novel antibacterial strategy for antibiotic resistance in helicobacter pylori. J. biomolecular structure dynamics, 1–16. doi: 10.1080/07391102.2023.2210674 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

