Atopic dermatitis-derived Staphylococcus aureus strains: what makes them special in the interplay with the host
- PMID: 37389215
- PMCID: PMC10303148
- DOI: 10.3389/fcimb.2023.1194254
Atopic dermatitis-derived Staphylococcus aureus strains: what makes them special in the interplay with the host
Abstract
Background: Atopic dermatitis (AD) is a chronic inflammatory skin condition whose pathogenesis involves genetic predisposition, epidermal barrier dysfunction, alterations in the immune responses and microbial dysbiosis. Clinical studies have shown a link between Staphylococcus aureus and the pathogenesis of AD, although the origins and genetic diversity of S. aureus colonizing patients with AD is poorly understood. The aim of the study was to investigate if specific clones might be associated with the disease.
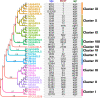
Methods: WGS analyses were performed on 38 S. aureus strains, deriving from AD patients and healthy carriers. Genotypes (i.e. MLST, spa-, agr- and SCCmec-typing), genomic content (e.g. virulome and resistome), and the pan-genome structure of strains have been investigated. Phenotypic analyses were performed to determine the antibiotic susceptibility, the biofilm production and the invasiveness within the investigated S. aureus population.
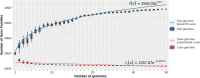
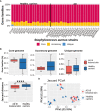
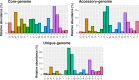
Results: Strains isolated from AD patients revealed a high degree of genetic heterogeneity and a shared set of virulence factors and antimicrobial resistance genes, suggesting that no genotype and genomic content are uniquely associated with AD. The same strains were characterized by a lower variability in terms of gene content, indicating that the inflammatory conditions could exert a selective pressure leading to the optimization of the gene repertoire. Furthermore, genes related to specific mechanisms, like post-translational modification, protein turnover and chaperones as well as intracellular trafficking, secretion and vesicular transport, were significantly more enriched in AD strains. Phenotypic analysis revealed that all of our AD strains were strong or moderate biofilm producers, while less than half showed invasive capabilities.
Conclusions: We conclude that in AD skin, the functional role played by S. aureus may depend on differential gene expression patterns and/or on post-translational modification mechanisms rather than being associated with peculiar genetic features.
Keywords: Staphylococcus aureus; antibiotic resistance; atopic dermatitis; biofilm; in silico genotype; pan-genome; whole-genome sequencing.
Copyright © 2023 Conte, Brunetti, Marazzato, Longhi, Maurizi, Raponi, Palamara, Grassi and Conte.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Alpdundar Bulut E., Bayyurt Kocabas B., Yazar V., Aykut G., Guler U., Salih B., et al. . (2020). Human gut commensal membrane vesicles modulate inflammation by generating M2-like macrophages and myeloid-derived suppressor cells. J. Immunol. 205, 2707–2718. doi: 10.4049/jimmunol.2000731 - DOI - PubMed
-
- Alsterholm M., Strömbeck L., Ljung A., Karami N., Widjestam J., Gillstedt M., et al. . (2017). Variation in Staphylococcus aureus colonization in relation to disease severity in adults with atopic dermatitis during a five-month follow-up. Acta Derm. Venereol. 97, 802–807. doi: 10.2340/00015555-2667 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

