Exploring the crop epigenome: a comparison of DNA methylation profiling techniques
- PMID: 37389288
- PMCID: PMC10306282
- DOI: 10.3389/fpls.2023.1181039
Exploring the crop epigenome: a comparison of DNA methylation profiling techniques
Abstract
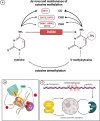
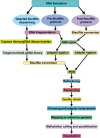
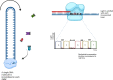
Epigenetic modifications play a vital role in the preservation of genome integrity and in the regulation of gene expression. DNA methylation, one of the key mechanisms of epigenetic control, impacts growth, development, stress response and adaptability of all organisms, including plants. The detection of DNA methylation marks is crucial for understanding the mechanisms underlying these processes and for developing strategies to improve productivity and stress resistance of crop plants. There are different methods for detecting plant DNA methylation, such as bisulfite sequencing, methylation-sensitive amplified polymorphism, genome-wide DNA methylation analysis, methylated DNA immunoprecipitation sequencing, reduced representation bisulfite sequencing, MS and immuno-based techniques. These profiling approaches vary in many aspects, including DNA input, resolution, genomic region coverage, and bioinformatics analysis. Selecting an appropriate methylation screening approach requires an understanding of all these techniques. This review provides an overview of DNA methylation profiling methods in crop plants, along with comparisons of the efficacy of these techniques between model and crop plants. The strengths and limitations of each methodological approach are outlined, and the importance of considering both technical and biological factors are highlighted. Additionally, methods for modulating DNA methylation in model and crop species are presented. Overall, this review will assist scientists in making informed decisions when selecting an appropriate DNA methylation profiling method.
Keywords: DNA methylation modulation; DNA methylation profiling; bisulfite sequencing; crop epigenome; immunological techniques; mass spectrometry; next-generation sequencing.
Copyright © 2023 Agius, Kapazoglou, Avramidou, Baranek, Carneros, Caro, Castiglione, Cicatelli, Radanovic, Ebejer, Gackowski, Guarino, Gulyás, Hidvégi, Hoenicka, Inácio, Johannes, Karalija, Lieberman-Lazarovich, Martinelli, Maury, Mladenov, Morais-Cecílio, Pecinka, Tani, Testillano, Todorov, Valledor and Vassileva.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
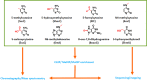
Figures
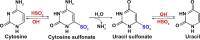


References
-
- Abid G., Mingeot D., Muhovski Y., Mergeai G., Aouida M., Abdelkarim S., et al. . (2017). Analysis of DNA methylation patterns associated with drought stress response in faba bean (Vicia faba l.) using methylation-sensitive amplification polymorphism (MSAP). Environ. Exp. Bot. 142, 34–44. doi: 10.1016/j.envexpbot.2017.08.004 - DOI
-
- Aliche E. B., Talsma W., Munnik T., Bouwmeester H. (2021). Characterization of maize root microbiome in two different soils by minimizing plant DNA contamination in metabarcoding analysis. Biol. Fertil. Soils 57, 731–737. doi: 10.1007/s00374-021-01555-3 - DOI
Publication types
LinkOut - more resources
Full Text Sources

