KG-Hub-building and exchanging biological knowledge graphs
- PMID: 37389415
- PMCID: PMC10336030
- DOI: 10.1093/bioinformatics/btad418
KG-Hub-building and exchanging biological knowledge graphs
Abstract
Motivation: Knowledge graphs (KGs) are a powerful approach for integrating heterogeneous data and making inferences in biology and many other domains, but a coherent solution for constructing, exchanging, and facilitating the downstream use of KGs is lacking.
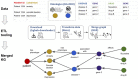
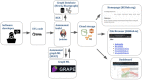
Results: Here we present KG-Hub, a platform that enables standardized construction, exchange, and reuse of KGs. Features include a simple, modular extract-transform-load pattern for producing graphs compliant with Biolink Model (a high-level data model for standardizing biological data), easy integration of any OBO (Open Biological and Biomedical Ontologies) ontology, cached downloads of upstream data sources, versioned and automatically updated builds with stable URLs, web-browsable storage of KG artifacts on cloud infrastructure, and easy reuse of transformed subgraphs across projects. Current KG-Hub projects span use cases including COVID-19 research, drug repurposing, microbial-environmental interactions, and rare disease research. KG-Hub is equipped with tooling to easily analyze and manipulate KGs. KG-Hub is also tightly integrated with graph machine learning (ML) tools which allow automated graph ML, including node embeddings and training of models for link prediction and node classification.
Availability and implementation: https://kghub.org.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures

References
-
- Auer S, Bizer C, Kobilarov G. et al. DBpedia: a nucleus for a web of open data. In: The Semantic Web. Berlin Heidelberg: Springer, 2007, 722–35.
-
- Bennett TD, Moffitt RA, Hajagos JG. et al. The national COVID cohort collaborative: clinical characterization and early severity prediction. medRxiv, 2021, preprint: not peer reviewed.
-
- Callahan TJ, Tripodi IJ, Hunter LE. et al. A framework for automated construction of heterogeneous Large-Scale biomedical knowledge graphs. bioRxiv, 2020, preprint: not peer reviewed.

