Bacterial DNA on the skin surface overrepresents the viable skin microbiome
- PMID: 37389570
- PMCID: PMC10328497
- DOI: 10.7554/eLife.87192
Bacterial DNA on the skin surface overrepresents the viable skin microbiome
Abstract
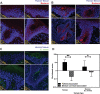
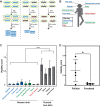
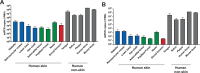
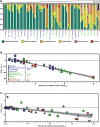
The skin microbiome provides vital contributions to human health. However, the spatial organization and viability of its bacterial components remain unclear. Here, we apply culturing, imaging, and molecular approaches to human and mouse skin samples, and find that the skin surface is colonized by fewer viable bacteria than predicted by bacterial DNA levels. Instead, viable skin-associated bacteria are predominantly located in hair follicles and other cutaneous invaginations. Furthermore, we show that the skin microbiome has a uniquely low fraction of viable bacteria compared to other human microbiome sites, indicating that most bacterial DNA on the skin surface is not associated with viable cells Additionally, a small number of bacterial families dominate each skin site and traditional sequencing methods overestimate both the richness and diversity of the skin microbiome. Finally, we performed an in vivo skin microbiome perturbation-recovery study using human volunteers. Bacterial 16S rRNA gene sequencing revealed that, while the skin microbiome is remarkably stable even in the wake of aggressive perturbation, repopulation of the skin surface is driven by the underlying viable population. Our findings help explain the dynamics of skin microbiome perturbation as bacterial DNA on the skin surface can be transiently perturbed but is replenished by a stable underlying viable population. These results address multiple outstanding questions in skin microbiome biology with significant implications for future efforts to study and manipulate it.
Keywords: Cutibacterium acnes; E. coli; Staphylococcus epidermidis; bacterial viability; human; infectious disease; microbiology; microbiome.
© 2023, Acosta et al.
Conflict of interest statement
EA, KL, BB, JL, XM, AP, MD, ZG No competing interests declared, DD Reviewing editor, eLife
Figures



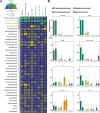
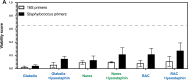
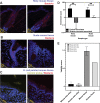
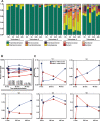


Update of
References
-
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted Oligonucleotide probes with flow Cytometry for analyzing mixed microbial populations. Applied and Environmental Microbiology. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. - DOI - PMC - PubMed
-
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet C, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope E, Da Silva R, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley G, Janssen S, Jarmusch AK, Jiang L, Kaehler B, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MG, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton J, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson, II MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CH, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG. QIIME 2: Reproducible, Interactive, Scalable, and Extensible Microbiome Data Science. PeerJ Preprints. 2018 doi: 10.7287/peerj.preprints.27295. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

