Autism-linked UBE3A gain-of-function mutation causes interneuron and behavioral phenotypes when inherited maternally or paternally in mice
- PMID: 37389991
- PMCID: PMC10530456
- DOI: 10.1016/j.celrep.2023.112706
Autism-linked UBE3A gain-of-function mutation causes interneuron and behavioral phenotypes when inherited maternally or paternally in mice
Abstract
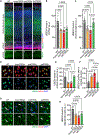
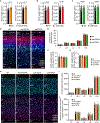
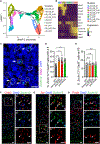
The E3 ubiquitin ligase Ube3a is biallelically expressed in neural progenitors and glial cells, suggesting that UBE3A gain-of-function mutations might cause neurodevelopmental disorders irrespective of parent of origin. Here, we engineered a mouse line that harbors an autism-linked UBE3AT485A (T503A in mouse) gain-of-function mutation and evaluated phenotypes in animals that inherited the mutant allele paternally, maternally, or from both parents. We find that paternally and maternally expressed UBE3AT503A results in elevated UBE3A activity in neural progenitors and glial cells. Expression of UBE3AT503A from the maternal allele, but not the paternal one, leads to a persistent elevation of UBE3A activity in neurons. Mutant mice display behavioral phenotypes that differ by parent of origin. Expression of UBE3AT503A, irrespective of its parent of origin, promotes transient embryonic expansion of Zcchc12 lineage interneurons. Phenotypes of Ube3aT503A mice are distinct from Angelman syndrome model mice. Our study has clinical implications for a growing number of disease-linked UBE3A gain-of-function mutations.
Keywords: Angelman syndrome; CP: Neuroscience; UBE3A; autism; neurodevelopmental disorders; single-cell RNA-seq.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
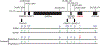


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

