A guide to the BRAIN Initiative Cell Census Network data ecosystem
- PMID: 37390046
- PMCID: PMC10313015
- DOI: 10.1371/journal.pbio.3002133
A guide to the BRAIN Initiative Cell Census Network data ecosystem
Abstract
Characterizing cellular diversity at different levels of biological organization and across data modalities is a prerequisite to understanding the function of cell types in the brain. Classification of neurons is also essential to manipulate cell types in controlled ways and to understand their variation and vulnerability in brain disorders. The BRAIN Initiative Cell Census Network (BICCN) is an integrated network of data-generating centers, data archives, and data standards developers, with the goal of systematic multimodal brain cell type profiling and characterization. Emphasis of the BICCN is on the whole mouse brain with demonstration of prototype feasibility for human and nonhuman primate (NHP) brains. Here, we provide a guide to the cellular and spatial approaches employed by the BICCN, and to accessing and using these data and extensive resources, including the BRAIN Cell Data Center (BCDC), which serves to manage and integrate data across the ecosystem. We illustrate the power of the BICCN data ecosystem through vignettes highlighting several BICCN analysis and visualization tools. Finally, we present emerging standards that have been developed or adopted toward Findable, Accessible, Interoperable, and Reusable (FAIR) neuroscience. The combined BICCN ecosystem provides a comprehensive resource for the exploration and analysis of cell types in the brain.
Copyright: © 2023 Hawrylycz et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: AR is a co-founder and equity holder of Celsius Therapeutics, an equity holder in Immunitas Therapeutics and, until 31 July 2020, was a scientific advisory board member of Thermo Fisher Scientific, Syros Pharmaceuticals, Asimov, and Neogene Therapeutics. From 1 August 2020, AR is an employee of Genentech and has equity in Roche. AR is a named inventor on multiple patents related to single cell and spatial genomics filed by or issued to the Broad Institute.
Figures

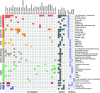
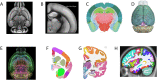
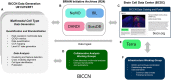



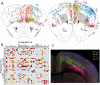
References
-
- RFA-MH-21-235: BRAIN Initiative Cell Atlas Network (BICAN): Comprehensive Center on Human and Non-human Primate Brain Cell Atlases (UM1 Clinical Trial Not Allowed) [Internet]. [cited 2022 Aug 18]. Available from: https://grants.nih.gov/grants/guide/rfa-files/RFA-MH-21-235.html.
Publication types
MeSH terms
Grants and funding
- RF1 MH123220/MH/NIMH NIH HHS/United States
- U19 MH114821/MH/NIMH NIH HHS/United States
- R24 MH114785/MH/NIMH NIH HHS/United States
- R01 MH113005/MH/NIMH NIH HHS/United States
- R24 MH117295/MH/NIMH NIH HHS/United States
- R01 NS039600/NS/NINDS NIH HHS/United States
- U19 MH114830/MH/NIMH NIH HHS/United States
- R01 NS086082/NS/NINDS NIH HHS/United States
- R01 MH094360/MH/NIMH NIH HHS/United States
- U01 MH114829/MH/NIMH NIH HHS/United States
- U01 MH114824/MH/NIMH NIH HHS/United States
- R01 NS096720/NS/NINDS NIH HHS/United States
- RF1 MH124605/MH/NIMH NIH HHS/United States
- U24 MH130988/MH/NIMH NIH HHS/United States
- RF1 MH128693/MH/NIMH NIH HHS/United States
- U01 MH117023/MH/NIMH NIH HHS/United States
- U19MH114824/MH/NIMH NIH HHS/United States
- R01MH123220/MH/NIMH NIH HHS/United States

