Synthetic Data Generation by Artificial Intelligence to Accelerate Research and Precision Medicine in Hematology
- PMID: 37390377
- PMCID: PMC10569771
- DOI: 10.1200/CCI.23.00021
Synthetic Data Generation by Artificial Intelligence to Accelerate Research and Precision Medicine in Hematology
Abstract
Purpose: Synthetic data are artificial data generated without including any real patient information by an algorithm trained to learn the characteristics of a real source data set and became widely used to accelerate research in life sciences. We aimed to (1) apply generative artificial intelligence to build synthetic data in different hematologic neoplasms; (2) develop a synthetic validation framework to assess data fidelity and privacy preservability; and (3) test the capability of synthetic data to accelerate clinical/translational research in hematology.
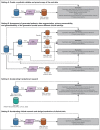
Methods: A conditional generative adversarial network architecture was implemented to generate synthetic data. Use cases were myelodysplastic syndromes (MDS) and AML: 7,133 patients were included. A fully explainable validation framework was created to assess fidelity and privacy preservability of synthetic data.
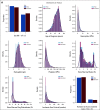
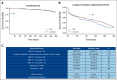
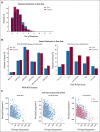
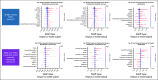
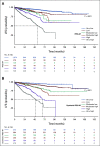
Results: We generated MDS/AML synthetic cohorts (including information on clinical features, genomics, treatment, and outcomes) with high fidelity and privacy performances. This technology allowed resolution of lack/incomplete information and data augmentation. We then assessed the potential value of synthetic data on accelerating research in hematology. Starting from 944 patients with MDS available since 2014, we generated a 300% augmented synthetic cohort and anticipated the development of molecular classification and molecular scoring system obtained many years later from 2,043 to 2,957 real patients, respectively. Moreover, starting from 187 MDS treated with luspatercept into a clinical trial, we generated a synthetic cohort that recapitulated all the clinical end points of the study. Finally, we developed a website to enable clinicians generating high-quality synthetic data from an existing biobank of real patients.
Conclusion: Synthetic data mimic real clinical-genomic features and outcomes, and anonymize patient information. The implementation of this technology allows to increase the scientific use and value of real data, thus accelerating precision medicine in hematology and the conduction of clinical trials.
Conflict of interest statement
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (
No other potential conflicts of interest were reported.
Figures










References
-
- Pencina MJ, Goldstein BA, D'Agostino RB: Prediction models—Development, evaluation, and clinical application. N Engl J Med 382:1583-1586, 2020 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

