Rapid Evolution of Glycan Recognition Receptors Reveals an Axis of Host-Microbe Arms Races beyond Canonical Protein-Protein Interfaces
- PMID: 37390614
- PMCID: PMC10329266
- DOI: 10.1093/gbe/evad119
Rapid Evolution of Glycan Recognition Receptors Reveals an Axis of Host-Microbe Arms Races beyond Canonical Protein-Protein Interfaces
Abstract
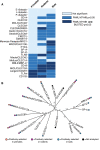
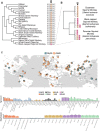
Detection of microbial pathogens is a primary function of many mammalian immune proteins. This is accomplished through the recognition of diverse microbial-produced macromolecules including proteins, nucleic acids, and carbohydrates. Pathogens subvert host defenses by rapidly changing these structures to avoid detection, placing strong selective pressures on host immune proteins that repeatedly adapt to remain effective. Signatures of rapid evolution have been identified in numerous immunity proteins involved in the detection of pathogenic protein substrates, but whether similar signals can be observed in host proteins engaged in interactions with other types of pathogen-derived molecules has received less attention. This focus on protein-protein interfaces has largely obscured the study of fungi as contributors to host-pathogen conflicts, despite their importance as a formidable class of vertebrate pathogens. Here, we provide evidence that mammalian immune receptors involved in the detection of microbial glycans have been subject to recurrent positive selection. We find that rapidly evolving sites in these genes cluster in key functional domains involved in carbohydrate recognition. Further, we identify convergent patterns of substitution and evidence for balancing selection in one particular gene, MelLec, which plays a critical role in controlling invasive fungal disease. Our results also highlight the power of evolutionary analyses to reveal uncharacterized interfaces of host-pathogen conflict by identifying genes, like CLEC12A, with strong signals of positive selection across mammalian lineages. These results suggest that the realm of interfaces shaped by host-microbe conflicts extends beyond the world of host-viral protein-protein interactions and into the world of microbial glycans and fungi.
Keywords: balancing selection; evolutionary conflict; host–pathogen interactions; microbial glycans; pattern recognition receptor; rapid evolution.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures


References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

