Effect of wash media type during PBMC isolation on downstream characterization of SARS-CoV-2-specific T cells
- PMID: 37390890
- PMCID: PMC10306416
- DOI: 10.1016/j.jim.2023.113520
Effect of wash media type during PBMC isolation on downstream characterization of SARS-CoV-2-specific T cells
Abstract
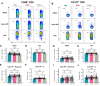
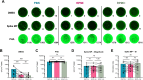
Protocols for the isolation of peripheral blood mononuclear cells (PBMCs) from whole blood vary greatly between laboratories, especially in published studies of SARS-CoV-2-specific T cell responses following infection and vaccination. Research on the effects of different wash media types or centrifugation speeds and brake usage during the PBMC isolation process on downstream T cell activation and functionality is limited. Blood samples from 26 COVID-19-vaccinated participants were processed with different PBMC isolation methods using either PBS or RPMI as the wash media with high centrifugation speed and brakes or RPMI as the wash media with low speed and brakes (RPMI+ method). SARS-CoV-2 spike-specific T cells were quantified and characterized via a flow cytometry-based activation induced markers (AIM) assay and an interferon-γ (IFNγ) FluoroSpot assay and responses were compared between processing methods. Samples washed with RPMI showed higher AIM+ CD4 T cell responses than those washed with PBS and showed a shift away from naïve and towards an effector memory phenotype. The activation marker OX40 showed higher SARS-CoV-2 spike-induced upregulation on RPMI-washed CD4 T cells, while differences in CD137 upregulation were minimal between processing methods. The magnitude of the AIM+ CD8 T cell response was similar between processing methods but showed higher stimulation indices. Background frequencies of CD69+ CD8 T cells were increased in PBS-washed samples and were associated with higher baseline numbers of IFNγ-producing cells in the FluoroSpot assay. Slower braking in the RPMI+ method did not improve detection of SARS-CoV-2-specific T cells and caused longer processing times. Thus, the use of RPMI media with full centrifugation brakes during the wash steps of PBMC isolation was found to be most effective and efficient. Further studies are needed to elucidate the pathways involved in RPMI-mediated preservation of downstream T cell activity.
Keywords: Centrifugation brakes; Flow cytometry; FluoroSpot; Processing; T cell; Wash media.
Copyright © 2023 Elsevier B.V. All rights reserved.
Figures



References
-
- Cytiva . 2020. HyClone Classical Media Formulation - RPMI-1640 Medium - Liquid Media.
-
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., Nakao C., Rayaprolu V., Rawlings S.A., Peters B., Krammer F., Simon V., Saphire E.O., Smith D.M., Weiskopf D., Sette A., Crotty S. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. - DOI - PMC - PubMed
-
- Gautam A., Donohue D., Hoke A., Miller S.A., Srinivasan S., Sowe B., Detwiler L., Lynch J., Levangie M., Hammamieh R., Jett M. Investigating gene expression profiles of whole blood and peripheral blood mononuclear cells using multiple collection and processing methods. PLoS One. 2019;14 doi: 10.1371/journal.pone.0225137. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

