Differentiation of IL-26+ TH17 intermediates into IL-17A producers via epithelial crosstalk in psoriasis
- PMID: 37391412
- PMCID: PMC10313793
- DOI: 10.1038/s41467-023-39484-4
Differentiation of IL-26+ TH17 intermediates into IL-17A producers via epithelial crosstalk in psoriasis
Abstract
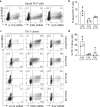
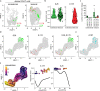
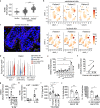
Interleukin (IL)-26 is a TH17 cytokine with known antimicrobial and pro-inflammatory functions. However, the precise role of IL-26 in the context of pathogenic TH17 responses is unknown. Here we identify a population of blood TH17 intermediates that produce high levels of IL-26 and differentiate into IL-17A-producing TH17 cells upon TGF-β1 exposure. By combining single cell RNA sequencing, TCR sequencing and spatial transcriptomics we show that this process occurs in psoriatic skin. In fact, IL-26+ TH17 intermediates infiltrating psoriatic skin induce TGF-β1 expression in basal keratinocytes and thereby promote their own differentiation into IL-17A-producing cells. Thus, our study identifies IL-26-producing cells as an early differentiation stage of TH17 cells that infiltrates psoriatic skin and controls its own maturation into IL17A-producing TH17 cells, via epithelial crosstalk involving paracrine production of TGF-β1.
© 2023. The Author(s).
Conflict of interest statement
J.D.D. and M.G. are inventors of a patent entitled “IL-26 inhibitors” (WO2017009392A1). R.G. declares ownership in Ozette Technologies. The remaining authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

