Analysis of polycomb repressive complex 2 (PRC2) subunits in Picea abies with a focus on embryo development
- PMID: 37391710
- PMCID: PMC10314529
- DOI: 10.1186/s12870-023-04359-9
Analysis of polycomb repressive complex 2 (PRC2) subunits in Picea abies with a focus on embryo development
Abstract
Background: Conserved polycomb repressive complex 2 (PRC2) mediates H3K27me3 to direct transcriptional repression and has a key role in cell fate determination and cell differentiation in both animals and plants. PRC2 subunits have undergone independent multiplication and functional divergence in higher plants. However, relevant information is still absent in gymnosperms.
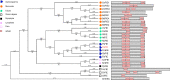
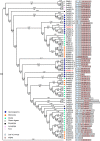
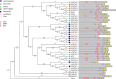
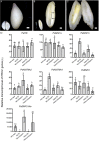
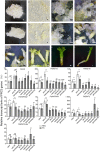
Results: To launch gymnosperm PRC2 research, we identified and cloned the PRC2 core component genes in the conifer model species Picea abies, including one Esc/FIE homolog PaFIE, two p55/MSI homologs PaMSI1a and PaMSI1b, two E(z) homologs PaKMT6A2 and PaKMT6A4, a Su(z)12 homolog PaEMF2 and a PaEMF2-like fragment. Phylogenetic and protein domain analyses were conducted. The Esc/FIE homologs were highly conserved in the land plant, except the monocots. The other gymnospermous PRC2 subunits underwent independent evolution with angiospermous species to different extents. The relative transcript levels of these genes were measured in endosperm and zygotic and somatic embryos at different developmental stages. The obtained results proposed the involvement of PaMSI1b and PaKMT6A4 in embryogenesis and PaKMT6A2 and PaEMF2 in the transition from embryos to seedlings. The PaEMF2-like fragment was predominantly expressed in the endosperm but not in the embryo. In addition, immunohistochemistry assay showed that H3K27me3 deposits were generally enriched at meristem regions during seed development in P. abies.
Conclusions: This study reports the first characterization of the PRC2 core component genes in the coniferous species P. abies. Our work may enable a deeper understanding of the cell reprogramming process during seed and embryo development and may guide further research on embryonic potential and development in conifers.
Keywords: H3K27me3; Norway spruce; Polycomb group; Seed development; Somatic embryogenesis.
© 2023. The Author(s).
Conflict of interest statement
We declare that we have no competing interest.
Figures



References
-
- Okano Y, Aono N, Hiwatashi Y, Murata T, Nishiyama T, Ishikawa T, Kubo M, Hasebe M. A polycomb repressive complex 2 gene regulates apogamy and gives evolutionary insights into early land plant evolution. Proc Natl Acad Sci U S A. 2009;106(38):16321–6. doi: 10.1073/pnas.0906997106. - DOI - PMC - PubMed
-
- Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci U S A. 2006;103(39):14631–6. doi: 10.1073/pnas.0606385103. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

