MRI-based breast cancer radiogenomics using RNA profiling: association with subtypes in a single-center prospective study
- PMID: 37391754
- PMCID: PMC10311893
- DOI: 10.1186/s13058-023-01668-7
MRI-based breast cancer radiogenomics using RNA profiling: association with subtypes in a single-center prospective study
Abstract
Background: There are few prospective studies on the correlations between MRI features and whole RNA-sequencing data in breast cancer according to molecular subtypes. The purpose of our study was to explore the association between genetic profiles and MRI phenotypes of breast cancer and to identify imaging markers that influences the prognosis and treatment according to subtypes.
Methods: From June 2017 to August 2018, MRIs of 95 women with invasive breast cancer were prospectively analyzed, using the breast imaging-reporting and data system and texture analysis. Whole RNA obtained from surgical specimens was analyzed using next-generation sequencing. The association between MRI features and gene expression profiles was analyzed in the entire tumor and subtypes. Gene networks, enriched functions, and canonical pathways were analyzed using Ingenuity Pathway Analysis. The P value for differential expression was obtained using a parametric F test comparing nested linear models and adjusted for multiple testing by reporting Q value.
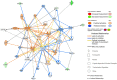
Results: In 95 participants (mean age, 53 years ± 11 [standard deviation]), mass lesion type was associated with upregulation of CCL3L1 (sevenfold) and irregular mass shape was associated with downregulation of MIR421 (sixfold). In estrogen receptor-positive cancer with mass lesion type, CCL3L1 (21-fold), SNHG12 (11-fold), and MIR206 (sevenfold) were upregulated, and MIR597 (265-fold), MIR126 (12-fold), and SOX17 (fivefold) were downregulated. In triple-negative breast cancer with increased standard deviation of texture analysis on precontrast T1-weighted imaging, CLEC3A (23-fold), SRGN (13-fold), HSPG2 (sevenfold), KMT2D (fivefold), and VMP1 (fivefold) were upregulated, and IGLC2 (73-fold) and PRDX4 (sevenfold) were downregulated (all, P < 0.05 and Q < 0.1). Gene network and functional analysis showed that mass type estrogen receptor-positive cancers were associated with cell growth, anti-estrogen resistance, and poor survival.
Conclusion: MRI characteristics are associated with the different expressions of genes related to metastasis, anti-drug resistance, and prognosis, depending on the molecular subtypes of breast cancer.
Keywords: Breast cancer; Magnetic resonance imaging; Molecular subtype; Radiogenomics; Texture analysis.
© 2023. The Author(s).
Conflict of interest statement
The authors have declared no competing interests.
Figures




References
-
- Grimm LJ, Zhang J, Mazurowski MA. Computational approach to radiogenomics of breast cancer: luminal A and luminal B molecular subtypes are associated with imaging features on routine breast MRI extracted using computer vision algorithms. J Magn Reson Imaging JMRI. 2015;42(4):902–907. - PubMed
-
- Leithner D, Horvat JV, Marino MA, Bernard-Davila B, Jochelson MS, Ochoa-Albiztegui RE, Martinez DF, Morris EA, Thakur S, Pinker K. Radiomic signatures with contrast-enhanced magnetic resonance imaging for the assessment of breast cancer receptor status and molecular subtypes: initial results. Breast Cancer Res BCR. 2019;21(1):106. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

