Exosome-derived circKIF20B suppresses gefitinib resistance and cell proliferation in non-small cell lung cancer
- PMID: 37394466
- PMCID: PMC10316567
- DOI: 10.1186/s12935-023-02974-y
Exosome-derived circKIF20B suppresses gefitinib resistance and cell proliferation in non-small cell lung cancer
Abstract
Background: The gefitinib resistance mechanism in non-small cell lung cancer (NSCLC) remains unclear, albeit exosomal circular RNA (circRNA) is known to possibly play a vital role in it.
Methods: We employed high-throughput sequencing techniques to detect the expressions of exosomal circRNA both in gefitinib-resistant and gefitinib-sensitive cells in this study. The circKIF20B expression was determined in serum exosomes and tissues of patients by qRT-PCR. The structure, stability, and intracellular localization of circKIF20B were verified by Sanger sequencing, Ribonuclease R (RNase R)/actinomycin D (ACTD) treatments, and Fluorescence in situ hybridization (FISH). The functions of circKIF20B were investigated by 5-Ethynyl-20-deoxyuridine (EdU), flow cytometry, Cell Counting Kit-8 (CCK-8), oxygen consumption rate (OCR), and xenograft model. Co-culture experiments were performed to explore the potential ability of exosomal circKIF20B in treating gefitinib resistance. The downstream targets of circKIF20B were determined by luciferase assay, RNA pulldown, and RNA immunoprecipitation (RIP).
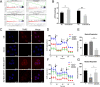
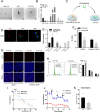
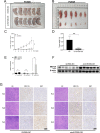
Results: We found that circKIF20B was poorly expressed in the serum exosomes of gefitinib-resistant patients (n = 24) and the tumor tissues of patients with NSCLC (n = 85). CircKIF20B was negatively correlated with tumor size and tumor stage. Decreasing circKIF20B was found to promote gefitinib resistance by accelerating the cell cycle, inhibiting apoptosis, and enhancing mitochondrial oxidative phosphorylation (OXPHOS), whereas increasing circKIF20B was found to restore gefitinib sensitivity. Mechanistically, circKIF20B is bound to miR-615-3p for regulating the MEF2A and then altering the cell cycle, apoptosis, and mitochondrial OXPHOS. Overexpressing circKIF20B parental cells can restore sensitivity to gefitinib in the recipient cells by upregulating the exosomal circKIF20B expression.
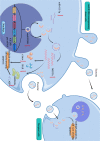
Conclusions: This study revealed a novel mechanism of circKIF20B/miR-615-3p/MEF2A signaling axis involving progression of gefitinib resistance in NSCLC. Exosomal circKIF20B is expected to be an easily accessible and alternative liquid biopsy candidate and potential therapeutic target in gefitinib-resistant NSCLC. The schematic diagram of mechanism in this study. Exosomal circKIF20B inhibits gefitinib resistance and cell proliferation by arresting the cell cycle, promoting apoptosis, and reducing OXPHOS via circKIF20B/miR-615-3p/MEF2A axis in NSCLC.
Keywords: Apoptosis; Cell cycle; CircKIF20B; Exosome; Gefitinib resistance.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Exosome-transmitted circVMP1 facilitates the progression and cisplatin resistance of non-small cell lung cancer by targeting miR-524-5p-METTL3/SOX2 axis.Drug Deliv. 2022 Dec;29(1):1257-1271. doi: 10.1080/10717544.2022.2057617. Drug Deliv. 2022. PMID: 35467477 Free PMC article.
-
Circ_0001658 regulates gefitinib resistance of non-small cell lung cancer through miR-409-3p/TWIST1 axis.Anticancer Drugs. 2022 Feb 1;33(2):158-166. doi: 10.1097/CAD.0000000000001257. Anticancer Drugs. 2022. PMID: 34694278
-
A novel circular RNA hsa_circRNA_103809/miR-377-3p/GOT1 pathway regulates cisplatin-resistance in non-small cell lung cancer (NSCLC).BMC Cancer. 2020 Dec 4;20(1):1190. doi: 10.1186/s12885-020-07680-w. BMC Cancer. 2020. PMID: 33276753 Free PMC article.
-
PLAUR Confers Resistance to Gefitinib Through EGFR/P-AKT/Survivin Signaling Pathway.Cell Physiol Biochem. 2018;47(5):1909-1924. doi: 10.1159/000491071. Epub 2018 Jun 29. Cell Physiol Biochem. 2018. PMID: 29961070
-
Identification of exosomal circRNA CD226 as a potent driver of nonsmall cell lung cancer through miR-1224-3p/high mobility group AT-hook 2 axis.Anticancer Drugs. 2022 Nov 1;33(10):1126-1138. doi: 10.1097/CAD.0000000000001357. Epub 2022 Aug 9. Anticancer Drugs. 2022. PMID: 35946568
Cited by
-
Circular RNAs in Cell Cycle Regulation of Cancers.Int J Mol Sci. 2024 May 31;25(11):6094. doi: 10.3390/ijms25116094. Int J Mol Sci. 2024. PMID: 38892280 Free PMC article. Review.
-
Harnessing EVs-ncRNA for Lung Cancer: From Oncogenic Pathways to Novel Diagnostic and Therapeutic Strategies.Int J Nanomedicine. 2025 Jul 14;20:9031-9054. doi: 10.2147/IJN.S528115. eCollection 2025. Int J Nanomedicine. 2025. PMID: 40689018 Free PMC article. Review.
-
Silencing of circRPPH1 Inhibits the Progression of Non-Small-Cell Lung Cancer Through miR-326/ERBB4 Signal Axis.Biochem Genet. 2025 Jun;63(3):2393-2408. doi: 10.1007/s10528-024-10824-3. Epub 2024 May 22. Biochem Genet. 2025. PMID: 38776052
-
Exosome therapeutics for non-small cell lung cancer tumorigenesis.Cancer Cell Int. 2024 Oct 30;24(1):360. doi: 10.1186/s12935-024-03544-6. Cancer Cell Int. 2024. PMID: 39478574 Free PMC article. Review.
-
The potential of targeting autophagy-related non-coding RNAs in the treatment of lung cancer.Front Pharmacol. 2025 May 14;16:1551258. doi: 10.3389/fphar.2025.1551258. eCollection 2025. Front Pharmacol. 2025. PMID: 40438586 Free PMC article. Review.
References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A, Cancer Statistics. 2021. CA Cancer J Clin. 2021;71(1):7–33. - PubMed
-
- Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Niksic M, et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–75. doi: 10.1016/S0140-6736(17)33326-3. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

