The diagnostic significance of the ZNF gene family in pancreatic cancer: a bioinformatics and experimental study
- PMID: 37396042
- PMCID: PMC10311482
- DOI: 10.3389/fgene.2023.1089023
The diagnostic significance of the ZNF gene family in pancreatic cancer: a bioinformatics and experimental study
Abstract
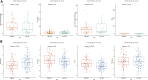
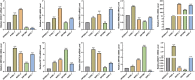
Background: Pancreatic adenocarcinoma (PAAD) is among the most devastating of all cancers with a poor survival rate. Therefore, we established a zinc finger (ZNF) protein-based prognostic prediction model for PAAD patients. Methods: The RNA-seq data for PAAD were downloaded from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) databases. Differentially expressed ZNF protein genes (DE-ZNFs) in PAAD and normal control tissues were screened using the "lemma" package in R. An optimal risk model and an independent prognostic value were established by univariate and multivariate Cox regression analyses. Survival analyses were performed to assess the prognostic ability of the model. Results: We constructed a ZNF family genes-related risk score model that is based on the 10 DE-ZNFs (ZNF185, PRKCI, RTP4, SERTAD2, DEF8, ZMAT1, SP110, U2AF1L4, CXXC1, and RMND5B). The risk score was found to be a significant independent prognostic factor for PAAD patients. Seven significantly differentially expressed immune cells were identified between the high- and low-risk patients. Then, based on the prognostic genes, we constructed a ceRNA regulatory network that includes 5 prognostic genes, 7 miRNAs and 35 lncRNAs. Expression analysis showed ZNF185, PRKCI and RTP4 were significantly upregulated, while ZMAT1 and CXXC1 were significantly downregulated in the PAAD samples in all TCGA - PAAD, GSE28735 and GSE15471 datasets. Moreover, the upregulation of RTP4, SERTAD2, and SP110 were verified by the cell experiments. Conclusion: We established and validated a novel, Zinc finger protein family - related prognostic risk model for patients with PAAD, that has the potential to inform patient management.
Keywords: TCGA; bioinformatics; pancreatic adenocarcinoma; prognostic risk model; zinc finger protein family.
Copyright © 2023 Zhu, Tu, Li, Li, Zhang, Jin, Li and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures











References
-
- Amedei A., Niccolai E., Benagiano M., Della Bella C., Cianchi F., Bechi P., et al. (2013). Ex vivo analysis of pancreatic cancer-infiltrating T lymphocytes reveals that ENO-specific Tregs accumulate in tumor tissue and inhibit Th1/Th17 effector cell functions. Cancer Immunol. Immunother. 62, 1249–1260. 10.1007/s00262-013-1429-3 - DOI - PMC - PubMed
-
- Chellappa S., Hugenschmidt H., Hagness M., Line P. D., Labori K. J., Wiedswang G., et al. (2016). Regulatory T cells that co-express RORγt and FOXP3 are pro-inflammatory and immunosuppressive and expand in human pancreatic cancer. Oncoimmunology 5, e1102828. 10.1080/2162402X.2015.1102828 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

