Construction and Identification of an NLR-Associated Prognostic Signature Revealing the Heterogeneous Immune Response in Skin Cutaneous Melanoma
- PMID: 37396711
- PMCID: PMC10312339
- DOI: 10.2147/CCID.S410723
Construction and Identification of an NLR-Associated Prognostic Signature Revealing the Heterogeneous Immune Response in Skin Cutaneous Melanoma
Abstract
Background: Skin cutaneous melanoma (SKCM) is the deadliest dermatology tumor. Ongoing researches have confirmed that the NOD-like receptors (NLRs) family are crucial in driving carcinogenesis. However, the function of NLRs signaling pathway-related genes in SKCM remains unclear.
Objective: To establish and identify an NLRs-related prognostic signature and to explore its predictive power for heterogeneous immune response in SKCM patients.
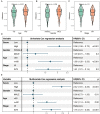
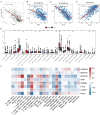
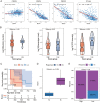
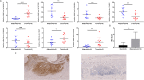
Methods: Establishment of the predictive signature using the NLRs-related genes by least absolute shrinkage and selection operator-Cox regression analysis (LASSO-COX algorithm). Through univariate and multivariate COX analyses, NLRs signature's independent predictive effectiveness was proven. CIBERSORT examined the comparative infiltration ratios of 22 distinct types of immune cells. RT-qPCR and immunohistochemistry implemented expression validation for critical NLRs-related prognostic genes in clinical samples.
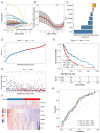
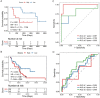
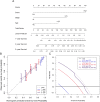
Results: The prognostic signature, including 7 genes, was obtained by the LASSO-Cox algorithm. In TCGA and validation cohorts, SKCM patients with higher risk scores had remarkably poorer overall survival. The independent predictive role of this signature was confirmed by multivariate Cox analysis. Additionally, a graphic nomogram demonstrated that the risk score of the NLRs signature has high predictive accuracy. SKCM patients in the low-risk group revealed a distinct immune microenvironment characterized by the significantly activated inflammatory response, interferon-α/γ response, and complement pathways. Indeed, several anti-tumor immune cell types were significantly accumulated in the low-risk group, including M1 macrophage, CD8 T cell, and activated NK cell. It is worth noting that our NLRs prognostic signature could serve as one of the promising biomarkers for predicting response rates to immune checkpoint blockade (ICB) therapy. Furthermore, the results of expression validation (RT-qPCR and IHC) were consistent with the previous analysis.
Conclusion: A promising NLRs signature with excellent predictive efficacy for SKCM was developed.
Keywords: NLR proteins; cutaneous melanoma; immune; prognosis; survival analysis.
© 2023 Geng et al.
Conflict of interest statement
The authors of this study declare that no conflicts of interest exist.
Figures


Similar articles
-
The fatty acid-related gene signature stratifies poor prognosis patients and characterizes TIME in cutaneous melanoma.J Cancer Res Clin Oncol. 2024 Jan 27;150(2):40. doi: 10.1007/s00432-023-05580-7. J Cancer Res Clin Oncol. 2024. PMID: 38279987 Free PMC article.
-
The Landscape of the Tumor Microenvironment in Skin Cutaneous Melanoma Reveals a Prognostic and Immunotherapeutically Relevant Gene Signature.Front Cell Dev Biol. 2021 Oct 1;9:739594. doi: 10.3389/fcell.2021.739594. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34660598 Free PMC article.
-
Analysis on tumor immune microenvironment and construction of a prognosis model for immune-related skin cutaneous melanoma.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2023 May 28;48(5):671-681. doi: 10.11817/j.issn.1672-7347.2023.230069. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2023. PMID: 37539569 Free PMC article. Chinese, English.
-
Identification of m6A-related lncRNAs-based signature for predicting the prognosis of patients with skin cutaneous melanoma.SLAS Technol. 2024 Feb;29(1):100101. doi: 10.1016/j.slast.2023.08.001. Epub 2023 Aug 2. SLAS Technol. 2024. PMID: 37541541
-
Development of an IFNγ response-related signature for predicting the survival of cutaneous melanoma.Cancer Med. 2020 Nov;9(21):8186-8201. doi: 10.1002/cam4.3438. Epub 2020 Sep 9. Cancer Med. 2020. PMID: 32902917 Free PMC article.
Cited by
-
Identification of an anoikis-related gene signature and characterization of immune infiltration in skin cutaneous melanoma.Medicine (Baltimore). 2024 Apr 26;103(17):e37900. doi: 10.1097/MD.0000000000037900. Medicine (Baltimore). 2024. PMID: 38669429 Free PMC article.
-
Natural Killer Cell Immune Checkpoints and Their Therapeutic Targeting in Cancer Treatment.Research (Wash D C). 2025 Jun 3;8:0723. doi: 10.34133/research.0723. eCollection 2025. Research (Wash D C). 2025. PMID: 40463500 Free PMC article.
-
Characterization of NOD-like receptor-based molecular heterogeneity in glioma and its association with immune micro-environment and metabolism reprogramming.Front Immunol. 2025 Jan 15;15:1498583. doi: 10.3389/fimmu.2024.1498583. eCollection 2024. Front Immunol. 2025. PMID: 39882240 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

