Promising AAV.U7snRNAs vectors targeting DMPK improve DM1 hallmarks in patient-derived cell lines
- PMID: 37397246
- PMCID: PMC10309041
- DOI: 10.3389/fcell.2023.1181040
Promising AAV.U7snRNAs vectors targeting DMPK improve DM1 hallmarks in patient-derived cell lines
Abstract
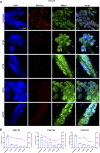
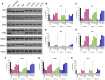
Myotonic dystrophy type 1 (DM1) is the most common form of muscular dystrophy in adults and affects mainly the skeletal muscle, heart, and brain. DM1 is caused by a CTG repeat expansion in the 3'UTR region of the DMPK gene that sequesters muscleblind-like proteins, blocking their splicing activity and forming nuclear RNA foci. Consequently, many genes have their splicing reversed to a fetal pattern. There is no treatment for DM1, but several approaches have been explored, including antisense oligonucleotides (ASOs) aiming to knock down DMPK expression or bind to the CTGs expansion. ASOs were shown to reduce RNA foci and restore the splicing pattern. However, ASOs have several limitations and although being safe treated DM1 patients did not demonstrate improvement in a human clinical trial. AAV-based gene therapies have the potential to overcome such limitations, providing longer and more stable expression of antisense sequences. In the present study, we designed different antisense sequences targeting exons 5 or 8 of DMPK and the CTG repeat tract aiming to knock down DMPK expression or promote steric hindrance, respectively. The antisense sequences were inserted in U7snRNAs, which were then vectorized in AAV8 particles. Patient-derived myoblasts treated with AAV8. U7snRNAs showed a significant reduction in the number of RNA foci and re-localization of muscle-blind protein. RNA-seq analysis revealed a global splicing correction in different patient-cell lines, without alteration in DMPK expression.
Keywords: U7snRNA; aav; gene therapy; myotonic dystrophy; spliceopathy.
Copyright © 2023 Almeida, Robriquet, Vetter, Huang, Neinast, Hernandez-Rosario, Rajakumar, Arnold, McBride, Flanigan, Weiss and Wein.
Conflict of interest statement
Since the performance of this work, Nationwide Children’s Hospital licensed the vector described herein to Audentes/Astellas Therapeutics. NW and KF have received royalty payments as a result of this license. NW is a Scientific Advisor for Alcyone Therapeutics. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Davis B. M., Mccurrach M. E., Taneja K. L., Singer R. H., Housman D. E. (1997). Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl. Acad. Sci. U. S. A. 94, 7388–7393. 10.1073/pnas.94.14.7388 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

