Maternal obesity alters the placental transcriptome in a fetal sex-dependent manner
- PMID: 37397247
- PMCID: PMC10309565
- DOI: 10.3389/fcell.2023.1178533
Maternal obesity alters the placental transcriptome in a fetal sex-dependent manner
Abstract
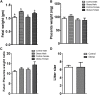
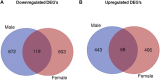
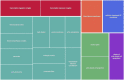
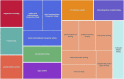
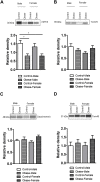
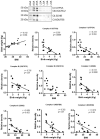
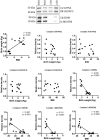
Infants born to obese mothers have an increased risk of developing obesity and metabolic diseases in childhood and adulthood. Although the molecular mechanisms linking maternal obesity during pregnancy to the development of metabolic diseases in offspring are poorly understood, evidence suggests that changes in the placental function may play a role. Using a mouse model of diet-induced obesity with fetal overgrowth, we performed RNA-seq analysis at embryonic day 18.5 to identify genes differentially expressed in the placentas of obese and normal-weight dams (controls). In male placentas, 511 genes were upregulated and 791 genes were downregulated in response to maternal obesity. In female placentas, 722 genes were downregulated and 474 genes were upregulated in response to maternal obesity. The top canonical pathway downregulated in maternal obesity in male placentas was oxidative phosphorylation. In contrast, sirtuin signaling, NF-kB signaling, phosphatidylinositol, and fatty acid degradation were upregulated. In female placentas, the top canonical pathways downregulated in maternal obesity were triacylglycerol biosynthesis, glycerophospholipid metabolism, and endocytosis. In contrast, bone morphogenetic protein, TNF, and MAPK signaling were upregulated in the female placentas of the obese group. In agreement with RNA-seq data, the expression of proteins associated with oxidative phosphorylation was downregulated in male but not female placentas of obese mice. Similarly, sex-specific changes in the protein expression of mitochondrial complexes were found in placentas collected from obese women delivering large-for-gestational-age (LGA) babies. In conclusion, maternal obesity with fetal overgrowth differentially regulates the placental transcriptome in male and female placentas, including genes involved in oxidative phosphorylation.
Keywords: fetal growth; gene expression; maternal–fetal exchange; mitochondria; trophoblast.
Copyright © 2023 Kelly, Chan, Powell, Cox, Jansson and Rosario.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources

