DeepCLEM: automated registration for correlative light and electron microscopy using deep learning
- PMID: 37397873
- PMCID: PMC10311120
- DOI: 10.12688/f1000research.27158.3
DeepCLEM: automated registration for correlative light and electron microscopy using deep learning
Abstract
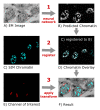
In correlative light and electron microscopy (CLEM), the fluorescent images must be registered to the EM images with high precision. Due to the different contrast of EM and fluorescence images, automated correlation-based alignment is not directly possible, and registration is often done by hand using a fluorescent stain, or semi-automatically with fiducial markers. We introduce "DeepCLEM", a fully automated CLEM registration workflow. A convolutional neural network predicts the fluorescent signal from the EM images, which is then automatically registered to the experimentally measured chromatin signal from the sample using correlation-based alignment. The complete workflow is available as a Fiji plugin and could in principle be adapted for other imaging modalities as well as for 3D stacks.
Keywords: Correlative Microscopy; Deep Learning; Image Registration; In-silico labeling.
Copyright: © 2023 Seifert R et al.
Conflict of interest statement
No competing interests were disclosed.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

