This is a preprint.
Fostering transparent medical image AI via an image-text foundation model grounded in medical literature
- PMID: 37398017
- PMCID: PMC10312868
- DOI: 10.1101/2023.06.07.23291119
Fostering transparent medical image AI via an image-text foundation model grounded in medical literature
Update in
-
Transparent medical image AI via an image-text foundation model grounded in medical literature.Nat Med. 2024 Apr;30(4):1154-1165. doi: 10.1038/s41591-024-02887-x. Epub 2024 Apr 16. Nat Med. 2024. PMID: 38627560
Abstract
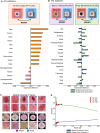
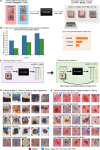
Building trustworthy and transparent image-based medical AI systems requires the ability to interrogate data and models at all stages of the development pipeline: from training models to post-deployment monitoring. Ideally, the data and associated AI systems could be described using terms already familiar to physicians, but this requires medical datasets densely annotated with semantically meaningful concepts. Here, we present a foundation model approach, named MONET (Medical cONcept rETriever), which learns how to connect medical images with text and generates dense concept annotations to enable tasks in AI transparency from model auditing to model interpretation. Dermatology provides a demanding use case for the versatility of MONET, due to the heterogeneity in diseases, skin tones, and imaging modalities. We trained MONET on the basis of 105,550 dermatological images paired with natural language descriptions from a large collection of medical literature. MONET can accurately annotate concepts across dermatology images as verified by board-certified dermatologists, outperforming supervised models built on previously concept-annotated dermatology datasets. We demonstrate how MONET enables AI transparency across the entire AI development pipeline from dataset auditing to model auditing to building inherently interpretable models.
Conflict of interest statement
Competing interests R.D. reports fees from L’Oreal, Frazier Healthcare Partners, Pfizer, DWA, and VisualDx for consulting; stock options from MDAcne and Revea for advisory board; and research funding from UCB.
Figures



References
-
- Daneshjou R., Yuksekgonul M., Cai Z. R., Novoa R. & Zou J. Y. SkinCon: A skin disease dataset densely annotated by domain experts for fine-grained debugging and analysis in Advances in Neural Information Processing Systems (eds Koyejo S. et al. ) 35 (Curran Associates, Inc., 2022), 18157–18167.
-
- Goel K., Gu A., Li Y. & Ré C. Model Patching: Closing the Subgroup Performance Gap with Data Augmentation in 9th International Conference on Learning Representations, ICLR 2021, Virtual Event, Austria, May 3-7, 2021 (OpenReview.net, 2021).
-
- Sagawa S., Koh P. W., Hashimoto T. B. & Liang P. Distributionally Robust Neural Networks in International Conference on Learning Representations (2020).
-
- Rajpurkar P. et al. MURA: Large Dataset for Abnormality Detection in Musculoskeletal Radiographs May 22, 2018. arXiv: 1712.06957[physics].
-
- Oakden-Rayner L., Dunnmon J., Carneiro G. & Re C. Hidden stratification causes clinically meaningful failures in machine learning for medical imaging in Proceedings of the ACM Conference on Health, Inference, and Learning ACM CHIL ‘20: ACM Conference on Health, Inference, and Learning (ACM, Toronto Ontario Canada, Apr. 2, 2020), 151–159. ISBN: 978-1-4503-7046-2. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
