This is a preprint.
Targeting Fibroblast-Endothelial Interactions in LAM Pathogenesis: 3D Spheroid and Spatial Transcriptomic Insights for Therapeutic Innovation
- PMID: 37398026
- PMCID: PMC10312665
- DOI: 10.1101/2023.06.12.544372
Targeting Fibroblast-Endothelial Interactions in LAM Pathogenesis: 3D Spheroid and Spatial Transcriptomic Insights for Therapeutic Innovation
Update in
-
Targeting fibroblast-endothelial cell interactions in LAM pathogenesis using 3D spheroid models and spatial transcriptomics.JCI Insight. 2025 Feb 4;10(6):e187899. doi: 10.1172/jci.insight.187899. JCI Insight. 2025. PMID: 39903528 Free PMC article.
Abstract
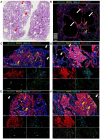
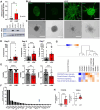
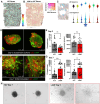
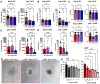
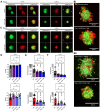
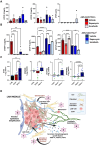
Lymphangioleiomyomatosis (LAM) is a progressive lung disease with limited treatments, largely due to an incomplete understanding of its pathogenesis. Lymphatic endothelial cells (LECs) invade LAM cell clusters, which include HMB-45-positive epithelioid cells and smooth muscle α-actin-expressing LAM-associated fibroblasts (LAMFs). Recent evidence shows that LAMFs resemble cancer-associated fibroblasts, with LAMF-LEC interactions contributing to disease progression. To explore these mechanisms, we used spatial transcriptomics on LAM lung tissues and identified a gene cluster enriched in kinase signaling pathways linked to myofibroblasts and co-expressed with LEC markers. Kinase arrays revealed elevated PDGFR and FGFR in LAMFs. Using a 3D co-culture spheroid model of primary LAMFs and LECs, we observed increased invasion in LAMF-LEC spheroids compared to non-LAM fibroblasts. Treatment with sorafenib, a multikinase inhibitor, significantly reduced invasion, outperforming Rapamycin. We also confirmed TSC2-null AML cells as key VEGF-A secretors, which was suppressed by sorafenib in both AML cells and LAMFs. These findings highlight VEGF-A and bFGF as potential therapeutic targets and suggest multikinase inhibition as a promising strategy for LAM.
Conflict of interest statement
Competing interests: Authors declare that they have no competing interests.
Figures


References
-
- Kitaichi M., Nishimura K., Itoh H., Izumi T., Pulmonary Lymphangioleiomyomatosis: a report of 46 patients including a clinicopathologic study of prognostic factors. Am J Respir Crit Care Med 151, 527–533 (1995). - PubMed
-
- Kwiatkowski D. J., Tuberous sclerosis: from tubers to mTOR. Ann Hum Genet 67, 87–96 (2003). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
