This is a preprint.
The debranching enzyme Dbr1 regulates lariat turnover and intron splicing
- PMID: 37398028
- PMCID: PMC10312976
- DOI: 10.21203/rs.3.rs-2931976/v1
The debranching enzyme Dbr1 regulates lariat turnover and intron splicing
Update in
-
The debranching enzyme Dbr1 regulates lariat turnover and intron splicing.Nat Commun. 2024 May 30;15(1):4617. doi: 10.1038/s41467-024-48696-1. Nat Commun. 2024. PMID: 38816363 Free PMC article.
Abstract
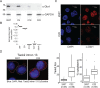
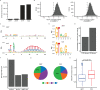
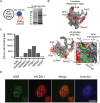
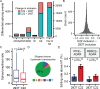
The majority of genic transcription is intronic. Introns are removed by splicing as branched lariat RNAs which require rapid recycling. The branch site is recognized during splicing catalysis and later debranched by Dbr1 in the rate-limiting step of lariat turnover. Through generation of the first viable DBR1 knockout cell line, we find the predominantly nuclear Dbr1 enzyme to encode the sole debranching activity in human cells. Dbr1 preferentially debranches substrates that contain canonical U2 binding motifs, suggesting that branchsites discovered through sequencing do not necessarily represent those favored by the spliceosome. We find that Dbr1 also exhibits specificity for particular 5' splice site sequences. We identify Dbr1 interactors through co-immunoprecipitation mass spectroscopy. We present a mechanistic model for Dbr1 recruitment to the branchpoint through the intron-binding protein AQR. In addition to a 20-fold increase in lariats, Dbr1 depletion increases exon skipping. Using ADAR fusions to timestamp lariats, we demonstrate a defect in spliceosome recycling. In the absence of Dbr1, spliceosomal components remain associated with the lariat for a longer period of time. As splicing is co-transcriptional, slower recycling increases the likelihood that downstream exons will be available for exon skipping.
Keywords: AQR; Dbr1; RNA processing; RNA splicing; intron lariats; lariat-seq; spliceosome.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests. We disclose W.G.F. as the founder of Walah Scientific and member of the scientific advisory board for Remix Therapeutics.
Figures

References
-
- Tseng C.-K. & Cheng S.-C. Both Catalytic Steps of Nuclear Pre-mRNA Splicing Are Reversible. Science 320, 1782–1784 (2008). - PubMed
-
- Ruskin B. & Green Michael R. An RNA Processing Activity That Debranches RNA Lariats. Science 229, 135–140 (1985). - PubMed
-
- Chapman K. B. & Boeke J. D. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell 65, 483–492 (1991). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

