This is a preprint.
De novo design of highly selective miniprotein inhibitors of integrins αvβ6 and αvβ8
- PMID: 37398153
- PMCID: PMC10312613
- DOI: 10.1101/2023.06.12.544624
De novo design of highly selective miniprotein inhibitors of integrins αvβ6 and αvβ8
Update in
-
De novo design of highly selective miniprotein inhibitors of integrins αvβ6 and αvβ8.Nat Commun. 2023 Sep 13;14(1):5660. doi: 10.1038/s41467-023-41272-z. Nat Commun. 2023. PMID: 37704610 Free PMC article.
Abstract
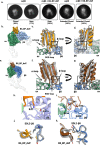
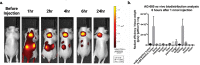
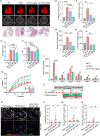
The RGD (Arg-Gly-Asp)-binding integrins αvβ6 and αvβ8 are clinically validated cancer and fibrosis targets of considerable therapeutic importance. Compounds that can discriminate between the two closely related integrin proteins and other RGD integrins, stabilize specific conformational states, and have sufficient stability enabling tissue restricted administration could have considerable therapeutic utility. Existing small molecules and antibody inhibitors do not have all of these properties, and hence there is a need for new approaches. Here we describe a method for computationally designing hyperstable RGD-containing miniproteins that are highly selective for a single RGD integrin heterodimer and conformational state, and use this strategy to design inhibitors of αvβ6 and αvβ8 with high selectivity. The αvβ6 and αvβ8 inhibitors have picomolar affinities for their targets, and >1000-fold selectivity over other RGD integrins. CryoEM structures are within 0.6-0.7Å root-mean-square deviation (RMSD) to the computational design models; the designed αvβ6 inhibitor and native ligand stabilize the open conformation in contrast to the therapeutic anti-αvβ6 antibody BG00011 that stabilizes the bent-closed conformation and caused on-target toxicity in patients with lung fibrosis, and the αvβ8 inhibitor maintains the constitutively fixed extended-closed αvβ8 conformation. In a mouse model of bleomycin-induced lung fibrosis, the αvβ6 inhibitor potently reduced fibrotic burden and improved overall lung mechanics when delivered via oropharyngeal administration mimicking inhalation, demonstrating the therapeutic potential of de novo designed integrin binding proteins with high selectivity.
Conflict of interest statement
Competing financial interests
A.R., L.S, X.D., J.L. T.S., D.B. are co-inventors on an International patent (Serial # PCT/US2020/057016) filed by University of Washington covering molecules and their uses described in this manuscript. C. O. is an employee of AstraZeneca and may own stock or stock options. M.G.C is an inventor on “Antibodies that bind integrin avb8 and uses thereof”, U.S. Patent US20210277125A1. A.R., H.B., J.C.K., M.S.S., M.C. and D.B. are inventors on a provisional patent describing the sequence and usage of αvβ8 integrins binders. A.R., L.S., J.C.K, H.B. and D.B. are co-founders of Lila Biologics and own stock or stock options in the company.
Figures


References
-
- Horan G. S. et al. Partial inhibition of integrin alpha(v)beta6 prevents pulmonary fibrosis without exacerbating inflammation. Am. J. Respir. Crit. Care Med. 177, 56–65 (2008). - PubMed
-
- Saini G. et al. αvβ6 integrin may be a potential prognostic biomarker in interstitial lung disease. Eur. Respir. J. 46, 486–494 (2015). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
