This is a preprint.
Mycobacterium tuberculosis resides in lysosome-poor monocyte-derived lung cells during chronic infection
- PMID: 37398178
- PMCID: PMC10312915
- DOI: 10.21203/rs.3.rs-3049913/v1
Mycobacterium tuberculosis resides in lysosome-poor monocyte-derived lung cells during chronic infection
Update in
-
Mycobacterium tuberculosis resides in lysosome-poor monocyte-derived lung cells during chronic infection.PLoS Pathog. 2024 May 3;20(5):e1012205. doi: 10.1371/journal.ppat.1012205. eCollection 2024 May. PLoS Pathog. 2024. PMID: 38701094 Free PMC article.
Abstract
Mycobacterium tuberculosis (Mtb) persists in lung myeloid cells during chronic infection. However, the mechanisms allowing Mtb to evade elimination are not fully understood. Here, we determined that in chronic phase, CD11clo monocyte-derived lung cells termed MNC1 (mononuclear cell subset 1), harbor more live Mtb than alveolar macrophages (AM), neutrophils, and less permissive CD11chi MNC2. Transcriptomic and functional studies of sorted cells revealed that the lysosome biogenesis pathway is underexpressed in MNC1, which have less lysosome content, acidification, and proteolytic activity than AM, and less nuclear TFEB, a master regulator of lysosome biogenesis. Mtb infection does not drive lysosome deficiency in MNC1. Instead, Mtb recruits MNC1 and MNC2 to the lungs for its spread from AM to these cells via its ESX-1 secretion system. The c-Abl tyrosine kinase inhibitor nilotinib activates TFEB and enhances lysosome function of primary macrophages and MNC1 and MNC2 in vivo, improving control of Mtb infection. Our results indicate that Mtb exploits lysosome-poor monocyte-derived cells for in vivo persistence, suggesting a potential target for host-directed tuberculosis therapy.
Conflict of interest statement
Competing interests The authors declare that they have no competing interests.
Figures




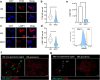

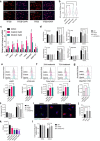
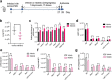
References
-
- Cegielski JP, Nahid P, Sotgiu G. The continued hunt for the elusive standard short regimen for treatment of multidrug-resistant tuberculosis. Eur Respir J 55, (2020). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

