This is a preprint.
CRISPR screens in iPSC-derived neurons reveal principles of tau proteostasis
- PMID: 37398204
- PMCID: PMC10312804
- DOI: 10.1101/2023.06.16.545386
CRISPR screens in iPSC-derived neurons reveal principles of tau proteostasis
Abstract
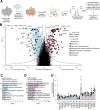
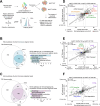
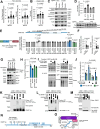
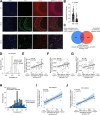
Aggregation of the protein tau defines tauopathies, which include Alzheimer's disease and frontotemporal dementia. Specific neuronal subtypes are selectively vulnerable to tau aggregation and subsequent dysfunction and death, but the underlying mechanisms are unknown. To systematically uncover the cellular factors controlling the accumulation of tau aggregates in human neurons, we conducted a genome-wide CRISPRi-based modifier screen in iPSC-derived neurons. The screen uncovered expected pathways, including autophagy, but also unexpected pathways, including UFMylation and GPI anchor synthesis. We discover that the E3 ubiquitin ligase CUL5SOCS4 is a potent modifier of tau levels in human neurons, ubiquitinates tau, and is a correlated with vulnerability to tauopathies in mouse and human. Disruption of mitochondrial function promotes proteasomal misprocessing of tau, which generates tau proteolytic fragments like those in disease and changes tau aggregation in vitro. These results reveal new principles of tau proteostasis in human neurons and pinpoint potential therapeutic targets for tauopathies.
Conflict of interest statement
DECLARATION OF INTERESTS M. K. is an inventor on US Patent 11,254,933 related to CRISPRi and CRISPRa screening, a co-scientific founder of Montara Therapeutics and serves on the Scientific Advisory Boards of Engine Biosciences, Alector, and Montara Therapeutics, and is an advisor to Modulo Bio and Recursion Therapeutics. The other authors declare no competing interests.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
