This is a preprint.
A non-enzymatic test for SARS-CoV-2 RNA using DNA nanoswitches
- PMID: 37398235
- PMCID: PMC10312858
- DOI: 10.1101/2023.05.31.23290613
A non-enzymatic test for SARS-CoV-2 RNA using DNA nanoswitches
Abstract
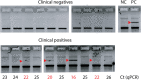
The emergence of a highly contagious novel coronavirus in 2019 led to an unprecedented need for large scale diagnostic testing. The associated challenges including reagent shortages, cost, deployment delays, and turnaround time have all highlighted the need for an alternative suite of low-cost tests. Here, we demonstrate a diagnostic test for SARS-CoV-2 RNA that provides direct detection of viral RNA and eliminates the need for costly enzymes. We employ DNA nanoswitches that respond to segments of the viral RNA by a change in shape that is readable by gel electrophoresis. A new multi-targeting approach samples 120 different viral regions to improve the limit of detection and provide robust detection of viral variants. We apply our approach to a cohort of clinical samples, positively identifying a subset of samples with high viral loads. Since our method directly detects multiple regions of viral RNA without amplification, it eliminates the risk of amplicon contamination and renders the method less susceptible to false positives. This new tool can benefit the COVID-19 pandemic and future emerging outbreaks, providing a third option between amplification-based RNA detection and protein antigen detection. Ultimately, we believe this tool can be adapted both for low-resource onsite testing as well as for monitoring viral loads in recovering patients.
Conflict of interest statement
Competing interests K.H. and W.P.W were joint inventors of DNA nanoswitches and hold several patents on the core technology. A.R.C., L.Z., D.Y., and C.H. are also inventors on nanoswitch related patents.
Figures



References
-
- WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int.
-
- Udugama B. et al. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 14, 3822–3835 (2020). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
