This is a preprint.
Mapping the genomic landscape of multidrug resistance in Plasmodium falciparum and its impact on parasite fitness
- PMID: 37398288
- PMCID: PMC10312498
- DOI: 10.1101/2023.06.02.543338
Mapping the genomic landscape of multidrug resistance in Plasmodium falciparum and its impact on parasite fitness
Update in
-
Mapping the genomic landscape of multidrug resistance in Plasmodium falciparum and its impact on parasite fitness.Sci Adv. 2023 Nov 10;9(45):eadi2364. doi: 10.1126/sciadv.adi2364. Epub 2023 Nov 8. Sci Adv. 2023. PMID: 37939186 Free PMC article.
Abstract
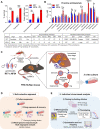
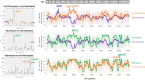
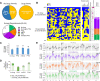
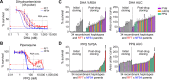
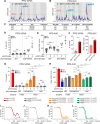
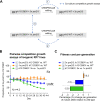
Drug-resistant Plasmodium falciparum parasites have swept across Southeast Asia and now threaten Africa. By implementing a P. falciparum genetic cross using humanized mice, we report the identification of key determinants of resistance to artemisinin (ART) and piperaquine (PPQ) in the dominant Asian KEL1/PLA1 lineage. We mapped k13 as the central mediator of ART resistance and identified secondary markers. Applying bulk segregant analysis, quantitative trait loci mapping and gene editing, our data reveal an epistatic interaction between mutant PfCRT and multicopy plasmepsins 2/3 in mediating high-grade PPQ resistance. Susceptibility and parasite fitness assays implicate PPQ as a driver of selection for KEL1/PLA1 parasites. Mutant PfCRT enhanced susceptibility to lumefantrine, the first-line partner drug in Africa, highlighting a potential benefit of opposing selective pressures with this drug and PPQ. We also identified that the ABCI3 transporter can operate in concert with PfCRT and plasmepsins 2/3 in mediating multigenic resistance to antimalarial agents.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures



References
-
- World Health Organization, WHO World Malaria Report 2022. https://www.who.int/teams/global-malaria-programme/reports/world-malaria..., (2022).
-
- Dondorp A. M., Nosten F., Yi P., Das D., Phyo A. P., Tarning J., Lwin K. M., Ariey F., Hanpithakpong W., Lee S. J., Ringwald P., Silamut K., Imwong M., Chotivanich K., Lim P., Herdman T., An S. S., Yeung S., Singhasivanon P., Day N. P., Lindegardh N., Socheat D., White N. J., Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361, 455–467 (2009). - PMC - PubMed
-
- Imwong M., Dhorda M., Myo Tun K., Thu A. M., Phyo A. P., Proux S., Suwannasin K., Kunasol C., Srisutham S., Duanguppama J., Vongpromek R., Promnarate C., Saejeng A., Khantikul N., Sugaram R., Thanapongpichat S., Sawangjaroen N., Sutawong K., Han K. T., Htut Y., Linn K., Win A. A., Hlaing T. M., van der Pluijm R. W., Mayxay M., Pongvongsa T., Phommasone K., Tripura R., Peto T. J., von Seidlein L., Nguon C., Lek D., Chan X. H. S., Rekol H., Leang R., Huch C., Kwiatkowski D. P., Miotto O., Ashley E. A., Kyaw M. P., Pukrittayakamee S., Day N. P. J., Dondorp A. M., Smithuis F. M., Nosten F. H., White N. J., Molecular epidemiology of resistance to antimalarial drugs in the Greater Mekong subregion: an observational study. Lancet Infect Dis 20, 1470–1480 (2020). - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
